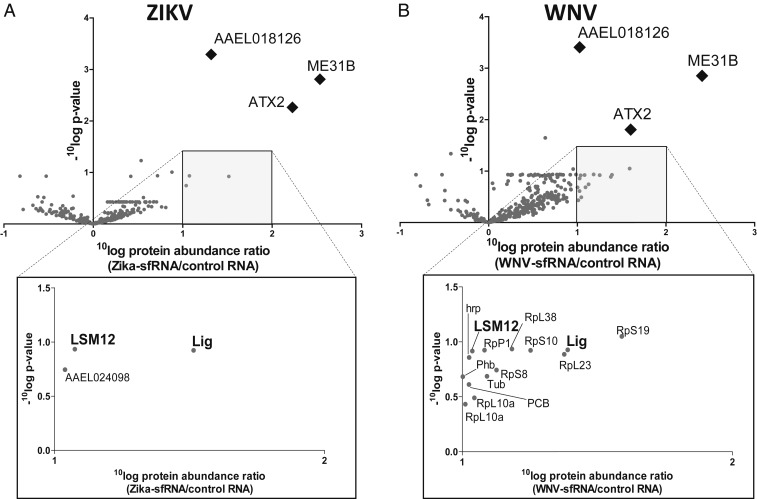
Fig. 3.
Identification of ZIKV and WNV sfRNA-binding proteins in Ae. aegypti cells using a streptavidin-binding aptamer approach and nano LC-MS/MS. 4XS1m-aptamer bound RNAs were bound to streptavidin beads and used to purify Ae. aegypti proteins with affinity for the bait RNA. Streptavidin beads were incubated with in vitro transcribed RNA of 4XS1m-ZIKV-sfRNA, 4XS1m-WNV-sfRNA, or 4XS1m-control. RNA-bound proteins were eluted by RNase A digestion of the bait RNA and identified and quantified by mass spectrometry. (A and B) Volcano plots of eluted proteins detected by mass spectrometry. The x axis shows the mean 10Log difference in protein abundance between the ZIKV sfRNA (A) or WNV sfRNA (B) and control samples from 3 independent biological replicates. The y axis shows the 10Log of the P value by Student’s t test from comparison of the protein abundance in the ZIKV sfRNA (A) or WNV sfRNA (B) with the control samples. Significantly enriched proteins are shown as diamonds. Insets below show proteins that are ≥10-fold enriched in the sfRNA samples but not observed significantly different from the control.