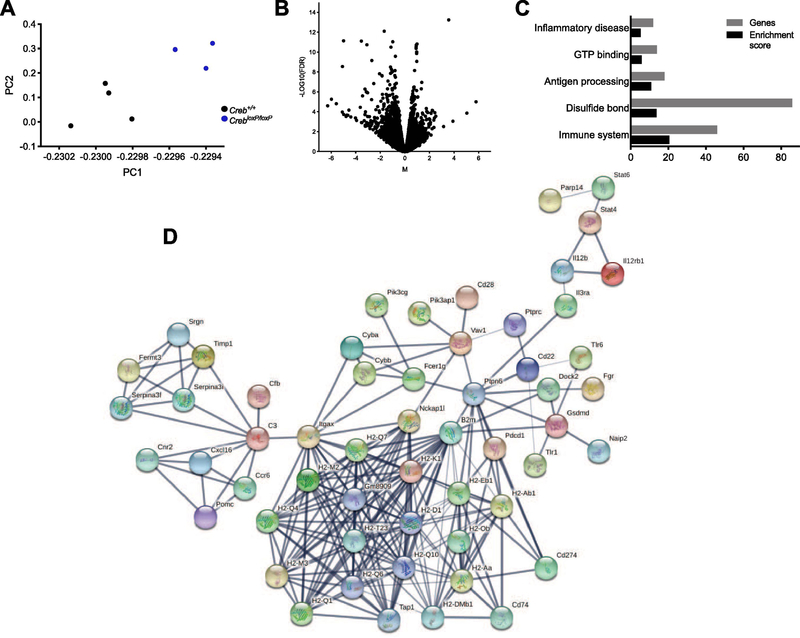
Figure 2. TRAP-sequencing was conducted on the hippocampus of CREB deletion and control mice.
A, nonlinear principle component analysis (PCA) of the complete RNA-seq data set found linear combinations of the 4 Creb+/+ and 3 CrebloxP/loxP samples. B, volcano plot from RNA-seq graphically shows the Log10FDR (False Discovery Rate) and the Fold Change (FC), CREB+/+FC-log2 CrebloxP/loxP / Creb+/+ FC. Genes with the largest fold change were downregulated. C, the top 205 downregulated genes were evaluated using DAVIDv6.8 for functional annotation clustering and listed according to enrichment score and the number of genes associated with the cluster. D, network analysis was conducted using STRING database. A network of 56 genes including cytokines, interleukins, and histocompatibility factors was identified.