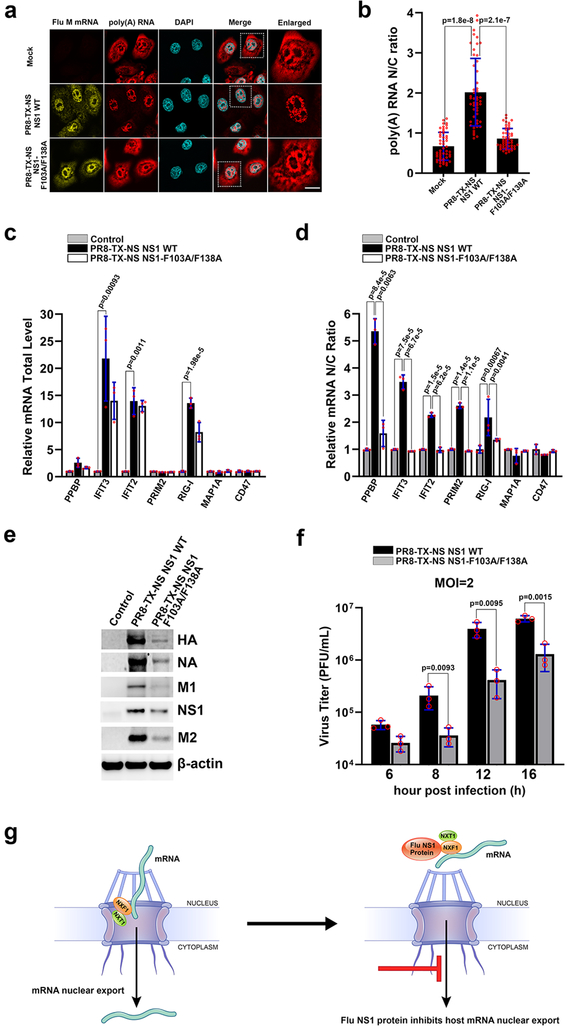
Fig. 4 |. Influenza virus mutant on the NXF1 binding site of NS1 allows nuclear export of host mRNAs and is attenuated.
a, HBECs were mock infected or ~ 100% infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A for 30h. Samples were subjected to RNA-FISH to detect viral M mRNA and poly(A) RNA. White square marks the enlarged panel on the right. Scale bar =10 μM. b, Quantification of fluorescence intensity in the nucleus and cytoplasm of samples in (a) followed by determination of nuclear to cytoplasmic ratios. Values are mean ± sd of 50 cells counted for each condition in three independent experiments. c, d, A549 cells were mock infected or infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A at MOI 2 for 6h. RNA was purified from total cell lysates (c) or from nuclear or cytoplasmic fractions (d). mRNAs selected based on transcriptome analysis shown in Supplementary Table 3 were quantified by qPCR. n=3. e, Cells were infected as in (c) except that infection lasted for 8h. Total cell lysates were subjected to western blot to detect the depicted proteins. n=3. f, Cells were infected as in (c) except that infection was performed for the indicated time points. Viral titers were measured in culture supernatants by plaque assay. n=3. The p-values (b, c, d, and f) were calculated using the unpaired, two-tailed Student’s t-test. g, Model for NS1-mediated inhibition of cellular mRNA nuclear export.