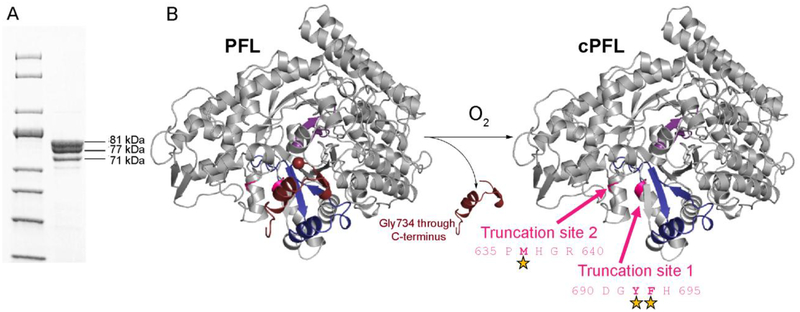
Figure 6:
cPFL is subject to proteolysis. A) SDS-PAGE of cPFL. Molecular weights of cPFL, 81 kDa, and two additional truncation products at 77 kDa (tlPFL) and 71 kDa (t2PFL) were extrapolated using a 10–200 kDa protein standard. These molecular weight bands correspond to the expected length of cPFL ending at residue 733, PFL truncated between residues 690–695 (tlPFL), and PFL truncated between 635–640 (t2PFL), respectively. B) Crystal structure of the PFL monomer showing residues 734–759 (end of C-terminus) in maroon, residues 695–733 in blue, the catalytic Cys loop in purple, and the rest of the monomer in gray. Upon exposure to oxygen, PFL is cleaved at Gly734. This cleavage exposes two truncation sites for E. coli chymotrypsin-like protease I activity. Positions of cleavage sites are indicated with stars.