Figure 1.
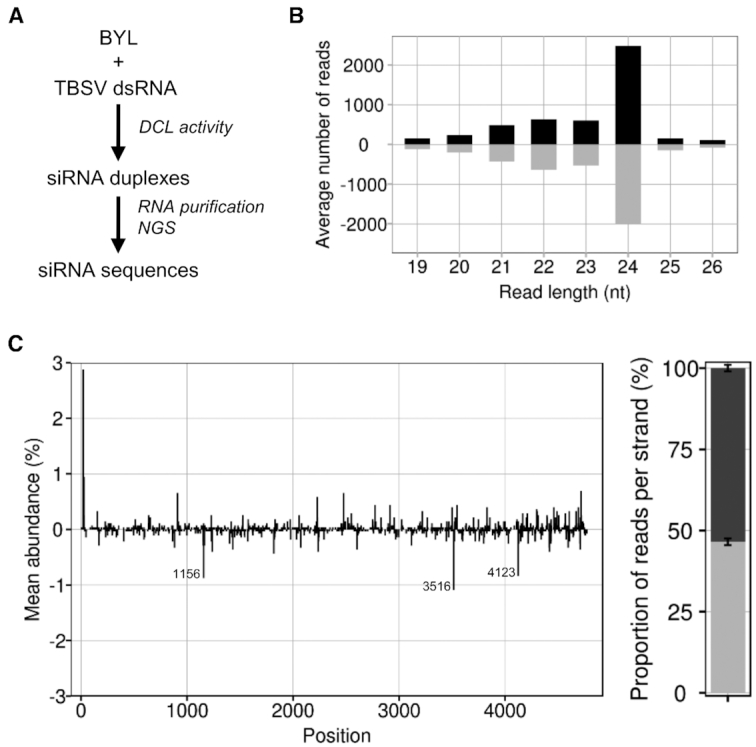
DCL-mediated generation of TBSV siRNAs in BYL. (A) Schematic representation of the in vitro ‘Dicer assay’ performed with double-stranded TBSV RNA. (B) Size distribution of sequenced TBSV siRNAs. Bars above the axis represent siRNAs derived from viral (+)RNA, bars below the axis represent siRNAs derived from viral (-)RNA. (C) Distribution and abundance of the sequenced 21 nt vsiRNAs aligned to the TBSV genome. Peaks above the axis represent siRNAs derived from viral (+)RNA, peaks below the axis represent siRNAs derived from viral (-)RNA. The peaks indicate the position of either the 5′ nucleotide of a vsiRNA with respect to the TBSV genome (in case of (+)vsiRNAs) or the TBSV genome position complementary to the 5′ nucleotide of a vsiRNA (in case of (-)vsiRNAs). The locations of the three most abundant (-)vsiRNA reads are specified. The chart on the right represents the relative abundance of the sum of (+) and (-)RNA derived siRNAs, respectively. Data represent mean ± S.E.M.
