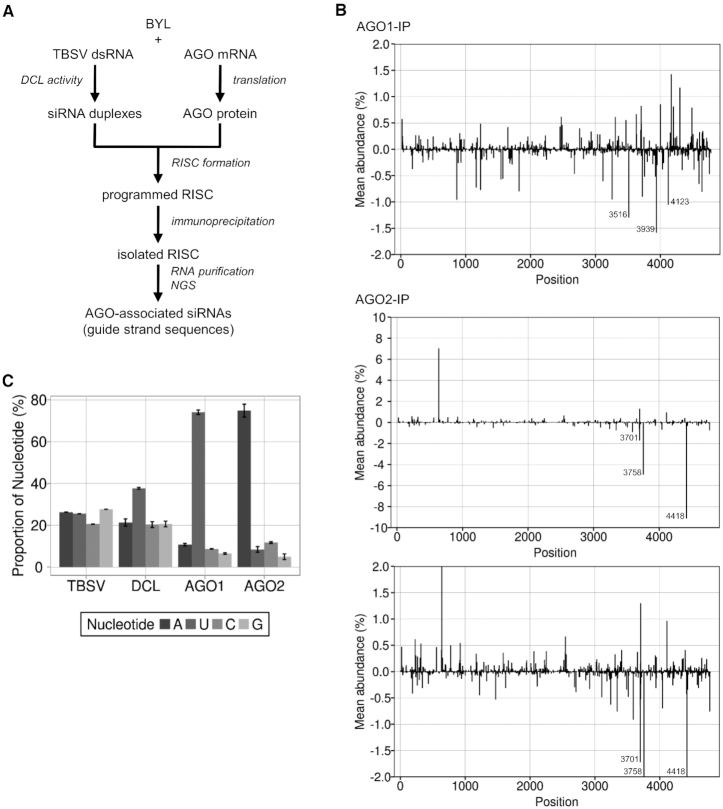
Figure 2.
Identification of RISC-incorporated TBSV siRNAs. (A) Schematic representation of the RISC immunoprecipitation procedure. FLAG-tagged AGO1 or AGO2 was generated by in vitro translation in the BYL in the presence of ds TBSV genomic RNA. The dsRNA was processed into vsiRNAs by the extract-endogenous DCLs and RISCs formed (‘programmed’) with these vsiRNAs. The RISCs were immunoprecipitated using an immobilized anti-FLAG antibody. Subsequently, siRNA guide strands were isolated and analyzed by NGS. (B) Distribution of sequenced 21 nt siRNA guide strands from immunoprecipitated AGO1/RISC or AGO2/RISC aligned to the TBSV genome. For better comparability, the additional image below shows a section of the AGO2 data with the same scaling as the AGO1 data. Peaks above the axis represent siRNAs derived from viral (+)RNA, peaks below the axis represent siRNAs derived from viral (-)RNA (see Figure 1 for the assignment of the peaks). The three most abundant (-)vsiRNA reads are shown. (C) Relative frequency of the respective 5′ terminal nucleotides of AGO1- and AGO2-associated 21 nt TBSV siRNA guide strands. The abundance was compared to the nucleotide composition of TBSV dsRNA and to the relative frequency of the 5′ terminal nucleotides of all 21 nt TBSV siRNAs that were generated by BYL-endogenous DCLs (see Figure 1). Data represent mean ± S.E.M.