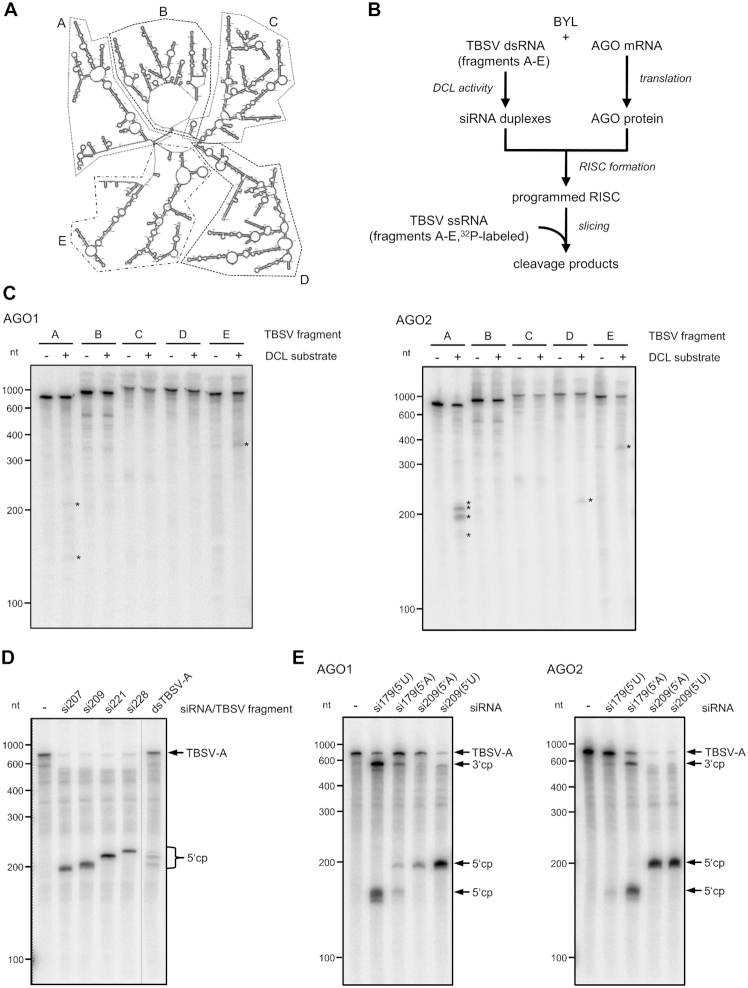
Figure 4.
Identification of vsiRNAs targeting particularly susceptible sites in the TBSV genome. (A) Secondary structure model of a TBSV RNA genome (47). Fragments A–E that were used in the slicer assay described below are indicated by dashed or spotted lines. (B) Schematic representation of the ‘slicer assay’ using a BYL-generated pool of TBSV siRNAs. Here, the AGO1 or AGO2/RISC was programmed with vsiRNAs that were produced from one of the ds TBSV fragments A–E by the BYL endogenous DCLs. As a control, the RISC-programming reaction was carried out in the absence of TBSV fragments. Subsequently, the corresponding 32P-labeled, single-stranded fragment transcript was added to the reaction and analyzed for cleavage by denaturing PAGE and autoradiography. (C) Results of the ‘slicer assay’ applying the BYL-generated vsiRNA pools. Detectable cleavage products are indicated by asterisks. (D) ‘Slicer assay’ performed with synthetic siRNAs that were selected as candidates for efficient cleavage of TBSV RNA fragment A. The assay was carried out as described in Figure 3 with AGO2. To assign the cleavage products (indicated as cp), a slicer assay containing the pool of ds fragment A-generated vsiRNAs (see B) was performed in parallel. (E) ‘Slicer assay’ performed with AGO1 or AGO2 and synthetic siRNAs 179 and 209 carrying different 5′ nucleotides. The assay was carried out with TBSV fragment A as target RNA as described in Figure 3. Cleavage products (cp) are indicated.