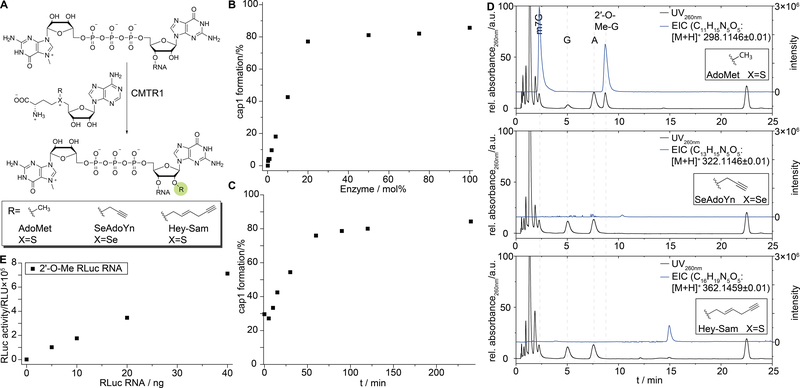
Figure 4.
A) Schematic representation of 2′-O modification by CMTR1 using AdoMet and AdoMet analogs SeAdoYn and AdoEnYn. B) Probing the catalytic efficiency of CMTR1160–549 by incubating the RNA substrate with varying ratios of enzyme. A short model RNA 7mGpppGAUC 8 (1 nmol) was incubated with varying amounts of CMTR160–549, a ten-fold excess of AdoMet and MTAN. Upon reaction, RNA was digested using snake venom phosphodiesterase and alkaline phosphatase and analyzed by RP-HPLC. C) Time course experiment of the in vitro 2′-O-methylation of 7mGpppGAUC by CMTR1160–549. Samples were taken after 0, 5, 10, 15, 30, 60, 90, 120 and 240 minutes and analyzed as described above. D) In vitro methylation assay of synthetic 7mGpppGAUC RNA 8 with CMTR1 using AdoMet, SeAdoYn or AdoEnYn as co-substrates. UV-chromatograms (260 nm, black traces) and the extracted ion-counts (EIC, blue traces) of the methylated (top panel), propargylated (middle panel) and hexenynylated (bottom panel) products are shown. E) In vitro translation experiment of Renilla luciferase RNA capped with the ARCA (3′-O-Me-7mGpppG) cap upon CMTR160–549 2′-O-methylation.