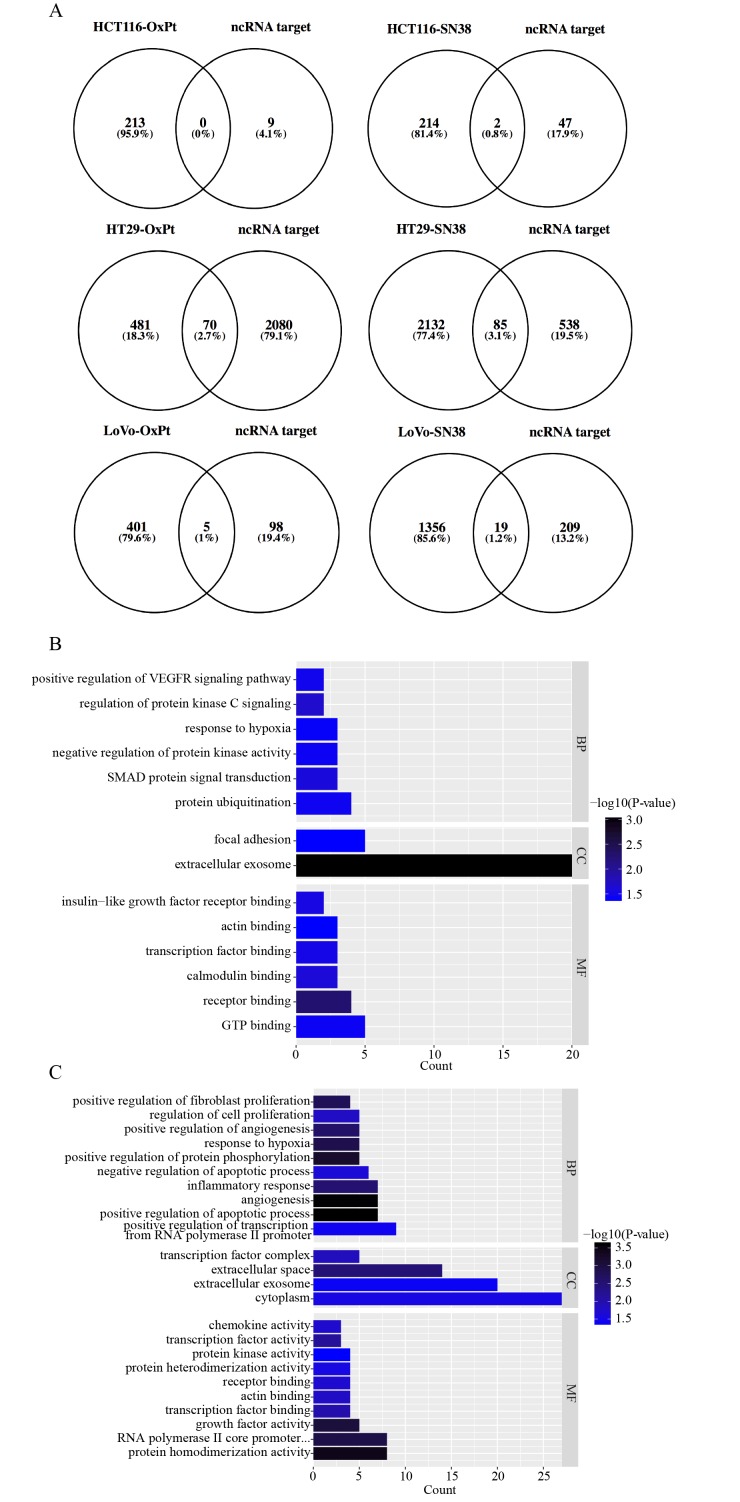
Figure 1.
Functional analyses of differentially expressed long non-coding RNA lncRNA-targeted DEGs. (A) A Venn diagram between lncRNA target genes in the database and screened DEGs for the six comparison groups. Histograms of GO functional classification of DEGs for the (B) OxPt resistance group and the (C) SN-38 resistance group. The x-axis represents the number of DEGs and the y-axis represents the GO terms. All GO terms were grouped into three ontologies: Biological process, cellular component and molecular function. The graph displays only significantly enriched GO terms (P<0.05); darker blue indicates greater significance. (D) A histogram of the KEGG pathway enrichment of DEGs screened in the SN-38 resistance group, where the x-axis represents the number of DEGs annotated in a pathway term; enriched pathways are shown on the y-axis. The graph displays only significantly enriched KEGG terms (P<0.05); darker red indicates greater significance. (E) A table listing biological process GO and KEGG terms in each group. lncRNA, long non-coding RNA; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed genes.