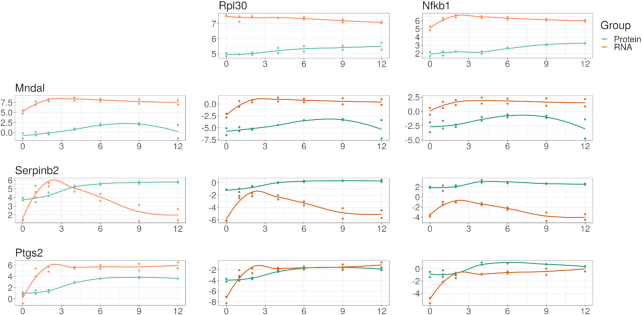
Figure 5:
mRNA abundance compared with newly synthesized protein abundance following LPS stimulation, illustrating the vertical integration of multi-omics data under a compositional framework. On the left margin, we show the log-abundance of 3 genes (MNDAL, SERPINB2, and PTGS2) as measured by RNA-Seq (orange) and mass spectrometry (blue). For compositional data, these abundances carry no meaning in isolation because the constrained total imposes a “closure bias.” On the top margin, we show the log-abundance of 2 references: RPL30 (chosen because its abundance is proportional to the geometric mean of the samples) and NFκB (chosen based on the hypothesis). In the middle, we show the abundance of the log-ratio of the left margin feature divided by the top margin reference (equivalent to left margin minus top margin in log space). MNDAL alone appears to exist more as mRNA than protein, which remains true when compared with both references. This suggests that MNDAL is translated with lower efficiency than RPL30 and NKκB. On the other hand, SERPINB2 alone appears to exist as mRNA and protein similarly on average; however, it actually exists more as protein than mRNA when compared with both references. This suggests that MNDAL is translated with greater efficiency than RPL30 and NKκB. PTGS2 alone appears to exist more as mRNA than protein, but this difference is less apparent when compared with both references. This suggests that PTGS2 is translated with a similar efficiency to RPL30 and NKκB. By choosing a reference shared between 2 multi-omics data sets, we can perform an analysis of vertically integrated data without any need for normalization.