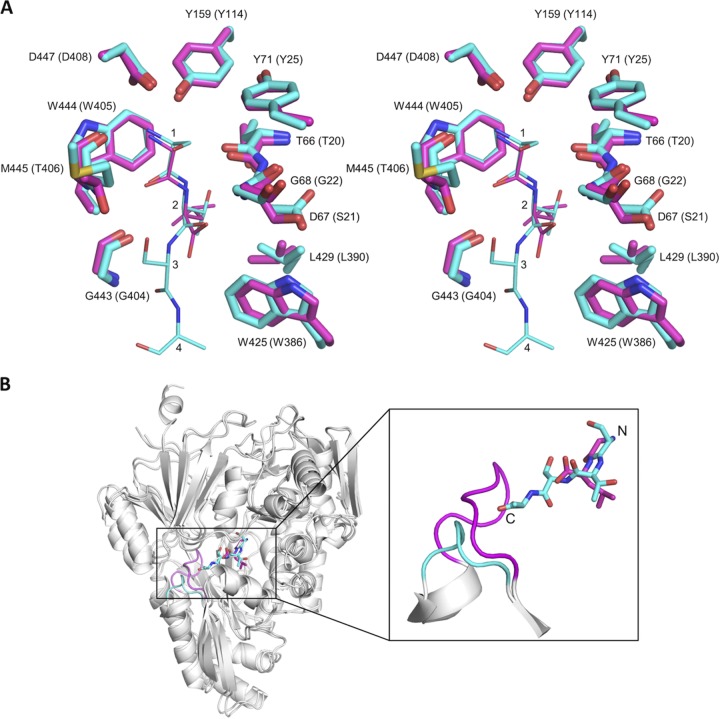
FIG 6.
Comparison of the structures of the HpDppA/STSA and EcDppA/GL crystal complexes. (A) Stereoview of the superposition of the ligand-binding sites of HpDppA (carbons colored cyan) and EcDppA (carbons colored magenta). Included are the tetrapeptide STSA bound to HpDppA and the dipeptide GL bound to EcDppA. The residues of the tetrapeptide ligand are labeled 1 to 4. Residue numbering in parentheses is for EcDppA. (B) Ribbon diagram of the HpDppA/STSA complex superimposed on the EcDppA/GL complex (PDB ID 1DPP), highlighting difference in lengths and positions of the loop from residue 397 to 401 in HpDppA (cyan) and the corresponding loop in EcDppA (magenta). The carbons in the tetrapeptide STSA bound to HpDppA and the dipeptide GL bound to EcDppA are also colored cyan and magenta, respectively.