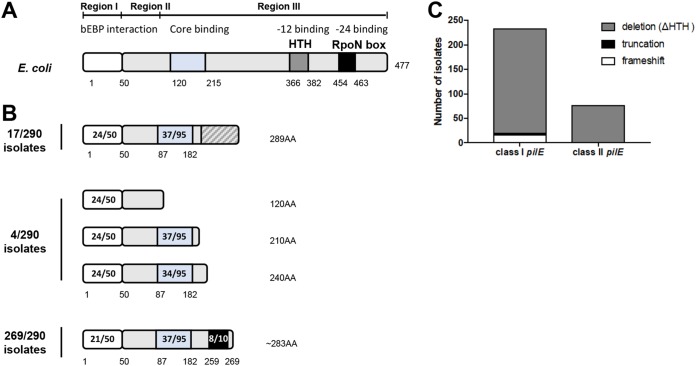
FIG 2.
Schematic diagrams of σN proteins in E. coli and N. meningitidis. (A) Schematic representation of the E. coli σN domain organization. Three regions have been defined: region I mediates interactions with activator proteins (bEBPs), region II is a linker with variable sequence, and region III comprises an RNAP core binding domain and two DNA binding domains, the HTH motif and RpoN box. (B) Schematic of the deduced domain organization of σN in N. meningitidis. Analysis of genomes of 290 meningococcal isolates revealed 34 different peptide sequences. Putative domains were identified based on EMBOSS Water pairwise sequence alignment with E. coli σN. Meningococcal σN was classified into three groups. σN proteins present in 17/290 isolates and 4/290 isolates harbor frameshift mutations or are truncated, respectively, and therefore lack any DNA binding domains. In the majority of isolates (269/290, 93%) σN lacks the HTH domain but retains an in-frame RpoN box. Amino acid numbers are shown. Numbers in the boxes indicate the numbers of identical amino acids compared to E. coli σN. (C) Distribution of putative σN types (frameshift, truncation, or deletion) in isolates with class I or class II pilE.