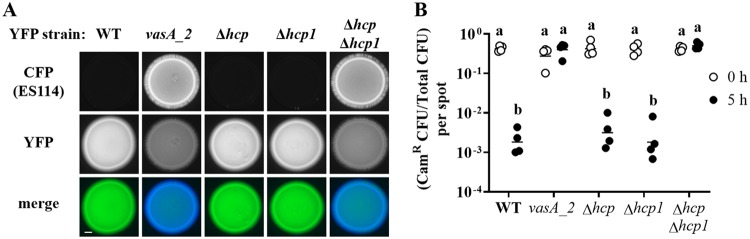
FIG 4.
Impact of hcp genes on the inhibition of ES114 growth by FQ-A001. (A) Coincubation assay with ES114 and the indicated FQ-A001-derived strains, harboring pYS112 and pSCV38, respectively. FQ-A001-derived strains are FQ-A001 (WT), ANS2098 (vasA_2), TIM416 (Δhcp), NPW57 (Δhcp1), and NPW58 (Δhcp Δhcp1). Shown are images of CFP fluorescence (top) or YFP fluorescence (center) and merged images of the bacterial growth resulting from the 24-h incubation of cellular mixtures spotted onto agar (bottom). Approximately 5 × 107 total CFU were introduced into each spot. Bar, 1 mm. (B) Ratio of Camr CFU to total CFU resulting from a 5-h coincubation of Camr-labeled ES114 with each of the indicated FQ-A001-derived strains. Each point represents the ratio associated with an individual spot at 0 h (open symbols) or 5 h (filled symbols), and each horizontal line represents the geometric mean (n = 4). Approximately 5 × 107 total CFU were introduced into each spot. Two-way analysis of variance revealed significant differences among the means of log-transformed ratios due to time (F1,30 = 54.78; P < 0.0001), genotype (F4,30 = 47.20; P < 0.0001), and their interaction (F4,30 = 54.78; P < 0.0001). A Sidak post hoc test was performed to compare the means statistically, with P values adjusted for multiple comparisons. Groups with different letters have significantly different medians (P < 0.0001), and groups with the same letter do not have significantly different medians (P ≥ 0.05).