Figure 7.
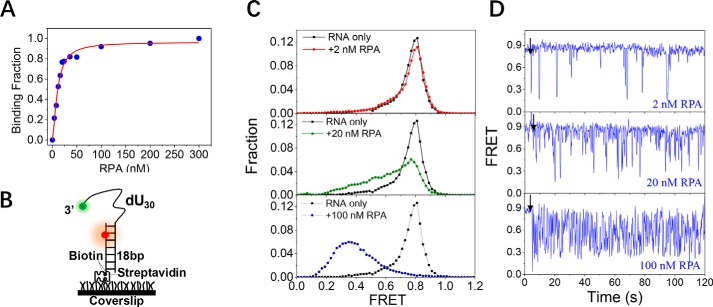
The binding of RPA to the single-stranded RNA is highly dynamic. A, binding fractions of RPA on the single-stranded RNA in 100 mm KCl were determined by fluorescence polarization assay. The binding curve was fitted by the Hill equation: y = [RPA]n/(KDn + [RPA]n) where y is the binding fraction, n is the Hill coefficient, and KD is the apparent dissociation constant. B, schematic representation of the smFRET experimental setup. C, FRET histograms of the RNA substrate dU30 alone and with various concentrations of RPA. Each FRET histogram was constructed from more than 300 traces. D, representative FRET traces of dU30 in the presence of RPA. Black arrows indicate the addition of RPA.