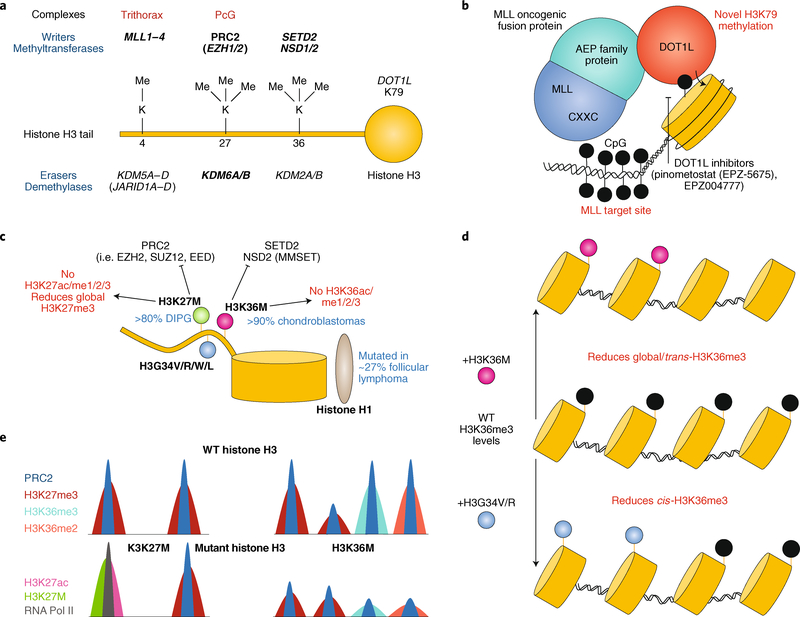
Fig. 3 |. Histone H3 methylation modifications and disruption in cancer.
a, Histone H3 methyltransferases and demethylases. Mutations to genes in bold are implicated in cancer. b, Depiction of MLL–ENL-rearranged leukaemia. The MLL CXXC domain targets the fusion protein to MLL target sites and the ENL domain recruits DOT1L methyltransferase activity, resulting in aberrant methylation. c, Schematic of oncohistone mutations in cancer and their antagonism with methyltransferases. d, The H3K36M mutation results in the global reduction of H3K36me3 levels, whereas the H3K34 mutation diminishes only cis-H3K36me3 levels. e, Schematic of H3K27M and H3K36M oncohistone chromatin occupancy compared to wild-type (WT) histone H3. H3K27M inhibits H3K27me3, resulting in RNA polymerase II (RNA Pol II) recruitment and activation, as assessed by H3K27ac levels. The H3K36M mutation reduces genome-wide H3K36me2/3 and H3K27me3 levels. DIPG, diffuse intrapontine glioma.