Abstract
Perturbations to mammalian SWI/SNF (mSWI/SNF) chromatin remodeling complexes have been widely implicated as driving events in cancer1. One such perturbation is the dual loss of the SMARCA4 and SMARCA2 ATPase subunits in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT)2–5, SMARCA4-deficient thoracic sarcomas6 and dedifferentiated endometrial carcinomas7. However, the consequences of dual ATPase subunit loss on mSWI/SNF complex subunit composition, chromatin targeting, DNA accessibility and gene expression remain unknown. Here we identify an ATPase module of subunits that is required for functional specification of BAF and PBAF subcomplexes. Using SMARCA4/2 ATPase mutant variants, we define the catalytic activity -dependent and -independent contributions of the ATPase module to the targeting of BAF and PBAF complexes on chromatin genome-wide. Finally, by linking distinct mSWI/SNF complex target sites to tumor-suppressive gene expression programs, we clarify the transcriptional consequences of SMARCA4/2 dual loss in SCCOHT.
Keywords: ATP-dependent chromatin remodeling, mammalian SWI/SNF (mSWI/SNF) complexes, chromatin regulation, gene expression, SCCOHT, ATPase, synthetic lethality, accessibility
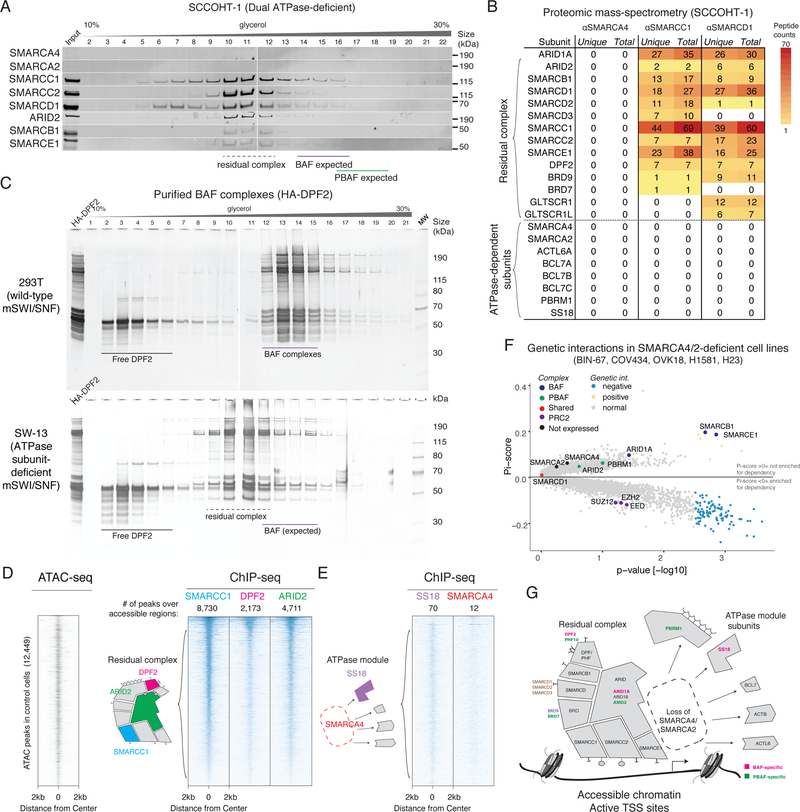
To define mSWI/SNF complex size and composition upon dual loss of the SMARCA4 and SMARCA2 ATPases, we leveraged two cell line models of SCCOHT: BIN-678,9 and SCCOHT-110, which we confirmed to be deficient for both SMARCA4 and SMARCA2 proteins (Supplementary Figure 1A). Using density-sedimentation of nuclear extracts coupled with immunoblot, we observed that a subset of remaining mSWI/SNF subunits co-sediment in the absence of SMARCA4 and SMARCA2 (Figure 1A, Supplementary Figure S1B), eluting in lower molecular-weight fractions than wild-type BAF and PBAF mSWI/SNF family subcomplexes (Supplementary Figure S1C). We confirmed that this co-sedimentation profile was a result of direct interaction between remaining subunits, and that residual complexes preserved mutually exclusive occupancy of subcomplex-specific subunits such as ARID1A (BAF-specific) and ARID2 (PBAF-specific) (Supplementary Figure 1D).
Figure 1.
Biochemical and genomic characterization of a residual complex in SMARCA4/SMARCA2 dual-deficient cancer cell lines.
A. Density sedimentation and immunoblot performed on SCCOHT-1 nuclear extracts. The residual mSWI/SNF complexes of both BAF and PBAF types exhibit similar elution profiles. See also Supplementary Figure 7a.
B. Peptides corresponding to mSWI/SNF subunits identified in immunoprecipitation/mass-spectrometry from SCCOHT-1 nuclear extract. SMARCC1 and SMARCD1 immunoprecipitations identify a cohort of subunits which remain stably bound to complexes in the absence of both ATPase subunits.
C. Density sedimentation performed on endogenously-purified BAF complexes from SMARCA4/2-intact (293T) and SMARCA4/2-deficient (SW-13) cell lines, using an HA-tagged DPF2 subunit as bait.
D. Heatmaps of chromatin accessibility (ATAC-seq) and residual mSWI/SNF occupancy (ChIP-seq) in the BIN-67 cell line, treated with a control Luciferase vector. Each row of the heatmap corresponds to a peak from the union of ATAC-seq and SMARCC1 ChIP-seq peaks in this condition. Rows are rank ordered by SMARCC1 occupancy. Each column shows the normalized read density for different experimental conditions across a 4-kb window centered at each peak.
E. Heatmaps reflecting SS18 and SMARCA4 ChIP-seq experiments performed in the BIN-67 cell line.
F. Genetic interaction data derived from CRISPR-Cas9-based screens performed in SMARCA4/2-dual-deficient and SMARCA4/A2-intact cell lines. Significance was calculated by a two-sided Wilcox test on the SMARCA4/2 deficient lines (n=5) against the WT lines (n=386). An FDR cutoff was 0.25 after BH correction was used.
G. Schematic of residual mSWI/SNF complex composition in the absence of the SMARCA4/2 ATPase subunits.
To comprehensively define the components of the residual complex, we performed large-scale immunoprecipitations from SCCOHT-1 nuclear extracts and subjected eluted proteins to mass-spectrometry-based proteomic analysis (Figure 1B, Supplementary Table 1). These studies defined two groups of mSWI/SNF subunits: residual complex components whose interactions were preserved in the absence of SMARCA4/2 ATPase subunits (ARID1A, ARID2, SMARCB1, SMARCC1/2, SMARCD1/2/3, SMARCE1, DPF2, GLTSCR1/L, and BRD7/9), or subunits that required SMARCA4/2 for complex binding (ACTL6A, BCL7A/B/C, PBRM1 and SS18), which we termed ‘ATPase module’ subunits. We replicated these results in the SMARCA4/2-deficient SW-13 cell line11, in which purified mSWI/SNF complexes similarly eluted at a lower molecular weight (Figure 1C) and selectively lacked ATPase module subunits (Supplementary Figure S1E), consistent with parallel studies from our laboratory12 as well as independent demonstrations that ACTL6A is destabilized upon loss of SMARCA4/2,13,14.
Given that the subunits of the residual complex contain numerous DNA-binding domains (such as the HMG domain of SMARCE1, winged helix domain of SMARCB1, etc.), we tested for residual genomic targeting in the BIN-67 cell line using chromatin immunoprecipitation followed by sequencing (ChIP-seq). Subunits in the residual complex (SMARCC1, DPF2, ARID2) bound accessible chromatin regions (Figure 1D) such as active transcriptional start sites (TSS) (Supplementary Figure 1F,G). By contrast, ChIP-seq performed on ATPase module subunits (SS18, SMARCA4) returned almost no significant peaks in these regions (Figure 1E), consistent with their absence from the complex and confirming the high specificity of mSWI/SNF ChIP-seq studies performed.
To test whether loss of SMARCA4/2 altered the essentiality of other mSWI/SNF subunits for cell proliferation, we performed genome-scale CRISPR-Cas9 fitness screening in the BIN-67 cell line15. Using a statistical framework to call genetic interactions from cancer cell line fitness screening data16, we found that mSWI/SNF complex members were significantly less essential in SMARCA4/2 dual- deficient cell lines (BIN-67 as well as COV434, OVK18, H1581, H23 cell lines) as compared to mSWI/SNF-intact cell lines (SMARCB1, p = 1.3e-3, SMARCE1, p = 1.0 e-3, Wilcox rank-sum test) (Figure 1F, Supplementary Figure 1H, Supplementary Table 2). We also recovered synthetic lethal relationships with PRC2 complex subunits, which have recently been reported17,18 (Figure 1F, Supplementary Figure 1H). Taken together, these results suggest that mSWI/SNF complexes exhibit a modular biochemical organization, and that the absence of the ATPase module causes BAF- and PBAF-specific subunits to co-elute as similarly-sized non-essential residual complexes that retain binding to accessible chromatin (Figure 1G).
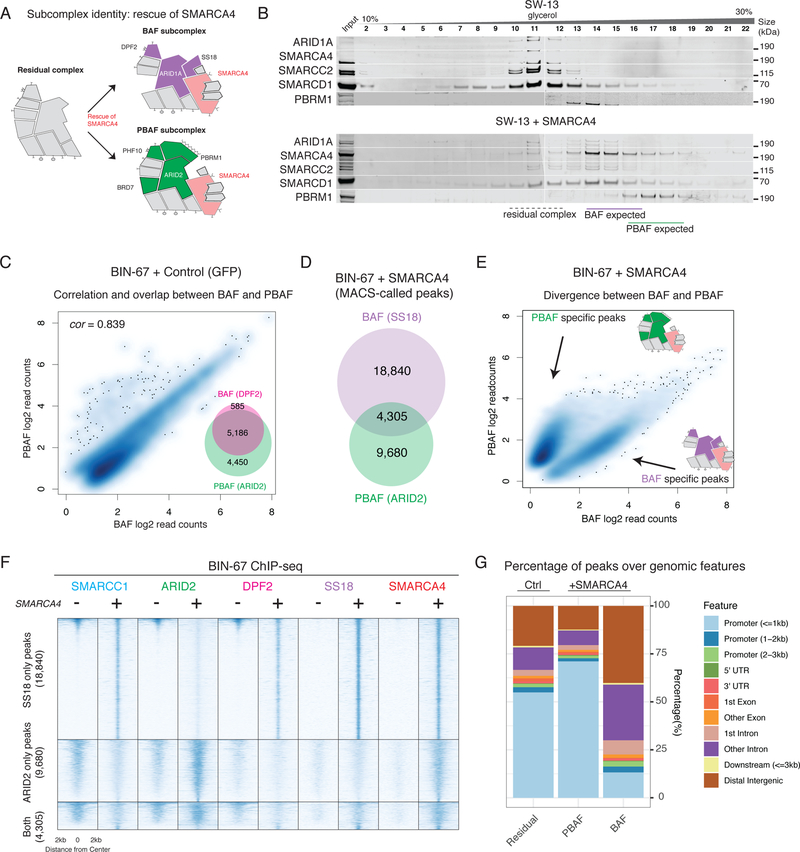
Given that the large majority (three quarters) of the subunits are common to both BAF and PBAF subcomplexes, the mechanisms by which mSWI/SNF subunits form functionally differentiated assemblies remain unknown. The modularity of the mSWI/SNF complex we identified suggested the possibility that the residual complex, once loaded with subcomplex-specific subunits, could attach to an ATPase module which contains either SS18 or PBRM1 (known to be mutually exclusive in BAF and PBAF complexes, respectively19,20), enabling functional specificity (Figure 2A). We confirmed that rescue of SMARCA4/2 in SW-13 cells sufficiently restored the protein-level abundance of ATPase module subunits (Supplementary Figure 2A) in a manner that was dose-dependent (Supplementary Figure 2B) and correlated with the overall abundance of SMARCA4 (correlation in immunofluorescence signal, Spearman ρ = 0.860, Supplementary Figure 2C). The presence of SMARCA4 also restored interactions between ATPase module subunits and residual complex components (Supplementary Figure 2D), including BAF-specific ARID1A-SS18 binding and PBAF-specific ARID2-PBRM1 binding (Supplementary Figure 2E–F).
Figure 2.
The ATPase module is required for BAF and PBAF subcomplex identity.
A. Schematic of BAF and PBAF subcomplexes and their formation upon rescue of SMARCA4 ATPase.
B. Density sedimentation experiments performed on nuclear extracts of SW-13 cells with and without SMARCA4 rescue. Expected fraction numbers for BAF and PBAF subcomplexes are indicated. See also Supplementary Figure 7b.
C. Log-scale scatterplot showing the normalized read counts from DPF2 and ARID2 ChIP-seq across a union set of DPF2 and ARID2 peaks (inset). Spearman correlation is shown.
D. Overlap between SS18 and ARID2 peaks in the SMARCA4 rescue condition in BIN-67 cells.
E. Log-scale scatterplot showing the normalized read counts from SS18 and ARID2 ChIP-seq across a union set of SS18 and ARID2 peaks upon SMARCA4 rescue.
F. Heatmaps reflecting ChIP-seq occupancy for SMARCC1, ARID2, DPF2, SS18, and SMARCA4 in control and SMARCA4 rescue conditions, split between in to SS18-only, ARID2-only, and shared occupancy. Rows are rank ordered by SMARCC1 occupancy in the control condition.
G. Distribution of mSWI/SNF peaks over genomic features in control and SMARCA4 rescue conditions.
Further, we found that BAF and PBAF complexes required the ATPase module for their distinct biochemical size profiles as well as their genomic targeting patterns on chromatin. Restoration of the ATPase module in SW-13 cells resulted in clear size-separation between BAF and PBAF complex subunits (Figure 2B), implying complete assembly of both complexes. In BIN-67 ATPase-deficient cells, genome-wide occupancies of BAF- and PBAF- specific subunits were strongly correlated (Spearman ρ = 0.839, Figure 2C), suggesting that the residual complexes lack the functional character of either BAF or PBAF despite the presence of complex-specific subunits. Restoration of the ATPase module increased chromatin affinity of both complexes (Supplementary Figure 2G) and led to gains in the number of significant mSWI/SNF peaks (Supplementary Figure 2H), which were predominantly non-overlapping between BAF and PBAF (Figure 2D–E), consistent with previous mSWI/SNF subunit reintroduction studies in cancer cells21 (although higher overlap at promoters has been observed in mSWI/SNF-intact cell lines22). These results indicate that integration of the ATPase module is required for BAF and PBAF complexes to target divergent genomic loci (Figure 2E–F), as demonstrated by the shift in PBAF localization toward TSSs and enrichment of BAF peaks distal to TSSs upon SMARCA4 rescue (Figure 2G).
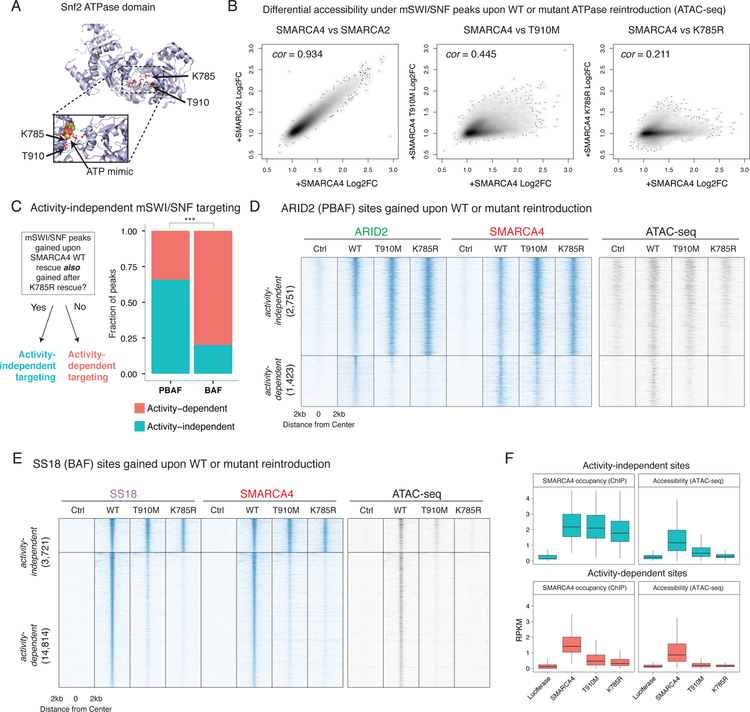
We next investigated how the ATPase module mediates divergent BAF and PBAF complex localization. Rescue of the ATPase subunit introduces two concurrent changes to mSWI/SNF complexes: (1) restored biochemical composition of the complex and (2) restored ATPase catalytic activity. To distinguish the relative contributions of these two events on BAF and PBAF localization and function, we rescued BIN-67 cells with either wild-type or ATPase mutant SMARCA4 (Figure 3A)23,24. SMARCA4 T910M and SMARCA4 K785R exhibited similar levels of protein expression as wild-type SMARCA4 (Supplementary Figure 3A) and recapitulated wild-type complex biochemical composition (Supplementary Figure 3B) as previously shown24,25, but exhibited either partial (T910M) or complete (K785R) losses of catalytic activity as assessed by in vitro ATPase assays (Supplementary Figure 3C) and genome-wide chromatin accessibility profiles upon rescue in BIN-67 cells (Figure 3B). Expression of either mutant variant resulted in partially reduced affinity of mSWI/SNF complexes to chromatin as compared to expression of wild-type SMARCA4 (Supplementary Figure 3D).
Figure 3.
Rescue of a catalytic activity-deficient ATPase module is sufficient to localize PBAF and BAF complexes to a subset of target sites genome-wide.
A. Mapping of T910M and K785R mutant residues to the ATP binding pocket of the yeast Swi2/Snf2 crystal structure (as performed in Hodges et al., 2018, NSMB) shows a proximity to a crystalized ATP mimic. Top: Overall structure of the Swi2 domain, mutated residues highlighted. Bottom: Enhanced view of ATP binding pocket in the presence of an ATP mimic.
B. Scatterplots of differential accessibility under SMARCC1 peaks upon rescue of different ATPase alleles. Each point in the scatterplot corresponds to a SMARCC1 peak called in the BIN-67 SMARCA4 rescue condition. Each axis displays the fold change in accessibility under each peak (as measured by ATAC-seq reads per million mapped to that peak) as compared to the Luciferase condition. Spearman correlation is shown.
C. Stacked bar plot of the proportion of BAF and PBAF peaks that are activity-dependent and activity-independent. Fisher’s two-sided exact test performed on total ChIP-seq peaks (n = 22,709), *** p < 1e−3
D. Heatmaps of chromatin accessibility and mSWI/SNF occupancy across ARID2 peaks gained upon rescue of a SMARCA4 allelic series. The ARID2 peakset is split into two groups: activity-independent peaks, and activity-dependent peaks. Rows are rank ordered by ARID2 occupancy in the wild type rescue condition.
E. Heatmaps of chromatin accessibility and mSWI/SNF occupancy across SS18 peaks that are gained upon rescue of a SMARCA4 allelic series. The SS18 peakset is split between activity-independent peaks, and activity-dependent peaks. Rows are rank ordered by SS18 occupancy in the wild type rescue condition.
F. Boxplots showing normalized readcounts across SS18 activity-independent (n=3,721) and activity-dependent peaks (n=14,814). First and third quartiles are denoted by lower and upper hinges. Whiskers extend to the smallest and largest values within 1.5x the interquartile range from the lower and upper hinges.
To assess whether gains in BAF and PBAF complex targeting can occur in the presence of the ATPase module but in the absence of wild-type levels of ATPase activity, we mapped the localization of mSWI/SNF complexes using ChIP-seq upon rescue of ATPase-dead SMARCA4 K785R (Figure 3C). Strong experimental agreement between SMARCA4 and SMARCC1 antibodies targeting mSWI/SNF complexes was detected in all reintroduction conditions (Supplementary Figure 3E). Unexpectedly, rescue with SMARCA4 K785R in BIN-67 cells was sufficient to target mSWI/SNF complexes to a subset of sites that are gained upon rescue of wild-type SMARCA4, and these sites mirrored the BAF- and PBAF-specific chromatin binding profiles observed with wild-type SMARCA4 rescue (Supplementary Figure 3F).
From this observation, we defined catalytic activity-dependent sites as those that were gained upon SMARCA4 WT rescue but not upon SMARCA4 K785R rescue, and catalytic activity-independent sites as those that were gained in both rescue conditions (Figure 3A). We further subdivided these peaks into BAF- and PBAF- specific sites by using the overlap between SMARCA4 and either SS18 (BAF) or ARID2 (PBAF) in the same condition, ensuring that each site represents high-confidence colocalization between at least two subunits of the same complex. We found that BAF and PBAF complexes exhibited significantly different proportions of catalytic activity-independent sites, with the majority of PBAF sites (~60%) but the minority of BAF sites (~20%) exhibiting catalytic activity-independent localization (Fisher’s test p < 2.2 e −16, Figure 3C). Over these BAF and PBAF activity-independent sites (Figure 3D–E), both the SMARCA4 K785R as well as the SMARCA4 T910M mutants exhibited comparable genomic occupancy compared to wild-type SMARCA4, although only the T910M mutant created partial chromatin accessibility under those sites due to its partial ATPase activity (Figure 3F). In contrast, at activity-dependent sites, only wild-type SMARCA4 was capable of generating mSWI/SNF de novo genomic occupancy and creating accessible sites (Figure 3F).
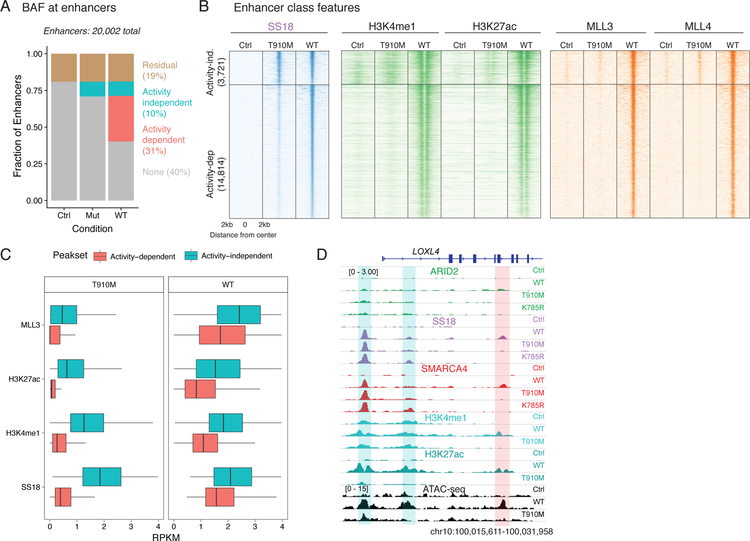
We then examined the role of these sites at BAF complex-occupied enhancers. We found that 10% of all enhancers in BIN-67 cells were targeted by BAF in a catalytic activity-independent manner and an additional ~30% in a catalytic activity-dependent manner (Figure 4A). We found that catalytically-deficient BAF complexes containing SMARCA4 T910M could target sites that were enriched for H3K4me1 and H3K27ac activating marks (Figure 4B), suggesting that the ATPase module directly or indirectly recognizes pre-established enhancer elements. However, catalytically-deficient BAF complexes could not target sites where these activating marks were absent (Figure 4B). These sites are neither heterochromatic nor silenced, as they lacked both the H3K9me3 and H3K27me3 marks, respectively (Supplementary Figure 4A). At these sites, wild-type ATPase activity was essential for targeting, and substantial increases in H3K4me1 and H3K27ac marks as well as recruitment of MLL3/4 methyltransferases (Figure 4C) occurred upon remodeling (ATAC-seq data shown in Figure 3E). In particular, enhancer activation at these sites likely requires full remodeling capability, as the partial ATPase activity of the T910M mutant (Figure 3B) was insufficient to trigger enhancer activation (Figure 4C,D) at these sites. These data suggest that previous models positing that H3K4me1 is required for BAF targeting are likely overly simplistic26, as BAF complex ATPase activity upon SMARCA4 rescue precedes the establishment of H3K4me1 by MLL3/4 in the majority of gained sites.
Figure 4.
Defining catalytic-dependent and -independent targeting of BAF complexes at enhancers.
A. Proportion of active enhancers occupied by BAF upon control and ATPase rescue.
B Heatmap showing each enhancer (ordered by SS18 occupancy) and the ChIP-seq profiles of H3K4me1, H3K27ac and MLL3/4.
C. Boxplot of ChIP-seq readcounts for MLL3, H3K27ac, H3K4me1 and SS18 epitopes over activity-independent (n=3,721) and -dependent (n=14,814) peaks across T910M and WT rescues. First and third quartiles are denoted by lower and upper hinges. Whiskers extend to the smallest and largest values within 1.5x the interquartile range from the lower and upper hinges.
D. Example browser track for the LOXL4. Activity-independent sites are highlighted in blue while activity-dependent sites are shown in red.
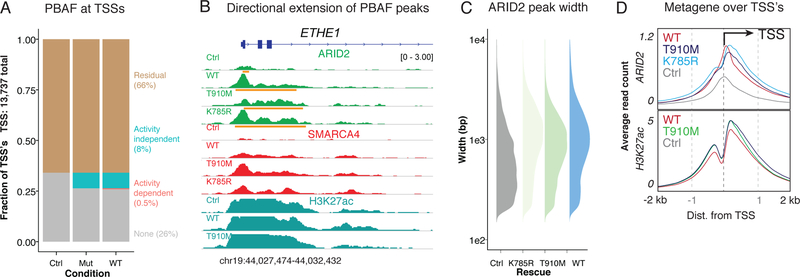
We also observed activity-independent effects for PBAF complexes upon rescue of the ATPase module. At PBAF-occupied TSS regions (within 4kb of TSS, Figure 5A), rescue of the ATPase module resulted in the directional extension of PBAF occupancy into gene bodies, for example, at the ETHE1 locus (Figure 5B). This directional extension resulted in marked increases in genome-wide PBAF peak width and occurred irrespective of whether the ATPase was catalytically active or catalytically dead (K785R) or deficient (T910M) (Figure 5C). Notably, the extent of PBAF spreading was concomitant with (and did not alter) the directional signatures of active transcription present at most promoters27, high H3K27Ac (Figure 5D) and depletion of H3K4me1 (Supplementary Figure 4B). A comparatively small percent (<1%, Figure 5A) of TSS regions encompassed activity-dependent sites (Supplementary Figure 4C). Taken together, these data suggest that the ATPase module on PBAF complexes enables recognition of transcriptional initiation signatures in a manner independent of catalytic activity.
Figure 5.
Directional PBAF complex occupancy along transcriptional initiation signatures requires the ATPase module, but not ATPase activity.
A. Proportion of active TSS’s occupied by PBAF (within 4 kb) upon control and ATPase rescue.
B. Example browser track for ETHE1, whose TSS is occupied by a narrow ARID2 peak in the control condition and wide ARID2 peaks upon ATPase subunit rescue. Broad peaks called by MACS 2.0 are shown in orange under ARID2 tracks.
C. Peak width distributions of ARID2 peaks called upon rescue of mutant and WT SMARCA4.
D. Metagene plot of ARID2 and H3K27ac occupancy over TSS’s.
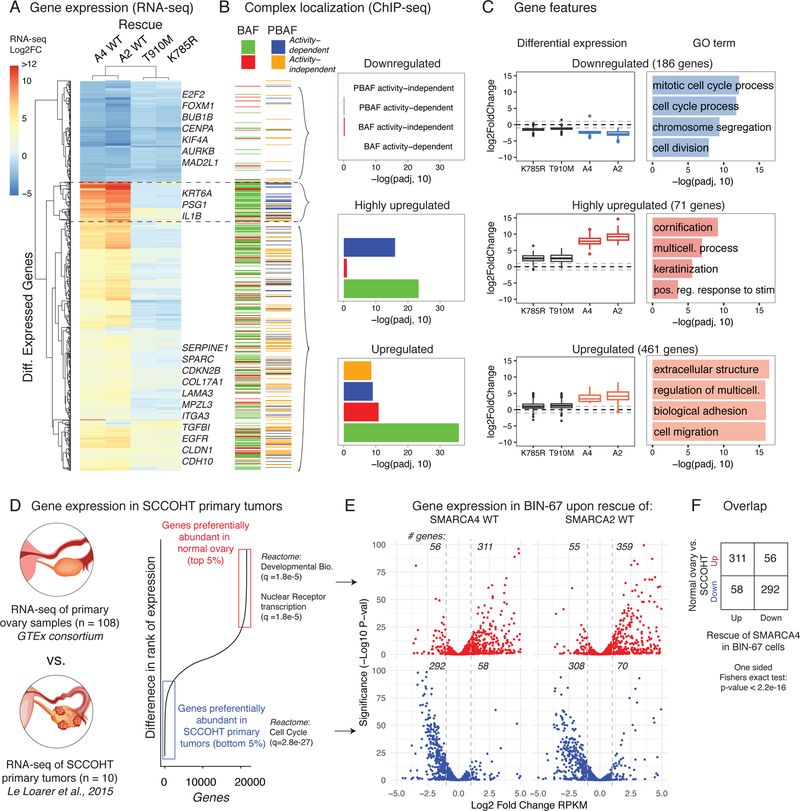
Next, to assess the gene regulatory effects of mSWI/SNF catalytic activity at these TSS and enhancer elements, we performed RNA-seq on BIN-67 cells in SMARCA4 wild-type and rescue conditions. In contrast to other enzymatic chromatin regulatory complexes such as MLL3/428, we found that the large majority of mSWI/SNF-driven gene expression changes in BIN-67 required mSWI/SNF enzymatic activity, with a ~7-fold increase in the number of upregulated genes identified between wild-type SMARCA4/2 and SMARCA4 mutant rescue conditions (Figure 6A, Supplementary Figure 5A, Supplementary Table 3). The downregulated, upregulated and highly upregulated gene sets as defined by hierarchical clustering differed in their requirements for direct targeting by mSWI/SNF family complexes. Downregulated genes, which were predominantly involved in cell cycle and chromosome segregation (i.e. BUB1B, CENPA, E2F2, FOXM1, etc), were not enriched for nearby BAF or PBAF complex localization relative to non-changing genes (Figure 6B), suggesting these gene changes were secondary downstream effects of SMARCA4 rescue. Highly upregulated genes involved in keratinization and other stimulus response genes (Figure 6C, Supplementary Figure 5B) were positively enriched for nearby BAF and PBAF complexes whose localization depended on their ATPase activity (Figure 6B), implying that these genes are targets of de novo enhancers created by both complexes. Finally, we found that PBAF and BAF catalytic activity- dependent and -independent targeting collaborated to upregulate genes in the third cluster (Figure 6B), which were involved in senescence and extracellular structure, cell migration and epithelial-mesenchymal transition (Figure 6C, Supplementary Figure 5C–D).
Figure 6.
Catalytically-active BAF and PBAF complexes collaborate to activate transcriptional programs that underlie differences between normal ovarian tissue and SCCOHT primary human tumors.
A. Heatmap of differential expression of the 700+ significantly changed genes upon SMARCA4 rescue, shown across all four rescue conditions in BIN-67 cells. Hierarchical clustering identifies three gene clusters, with representative genes labeled.
B. (Left) Heatmap showing the mSWI/SNF chromatin occupancy status of the genes in (A). BAF and PBAF occupancies are shown separately, and activity-independent and dependent peaks are indicated. (Right) Enrichment of each of the four peak groups in the three gene clusters using a one-sided Fisher’s Exact test, corrected for multiple hypotheses with the BH correction (Downregulated: n = 186; Highly upregulated: n = 71; Upregulated: n = 461).
C. (Left) Boxplots showing differential gene expression across each of the three clusters. First and third quartiles are denoted by lower and upper hinges. Whiskers extend to the smallest and largest values within 1.5x the interquartile range (IQR) from the lower and upper hinges. Outliers beyond 1.5x IQR are plotted as individual points. (Right) Top four GO enrichments for genes in these three clusters. Enrichment was calculated by hypergeometric test after BH false discovery rate correction by the Gorilla package.
D. RNA-seq data from SCCOHT patient tumors were compared to primary ovarian samples from the GTEx consortium to generate two gene sets: genes that were preferentially abundant in normal ovarian tissue (red) and preferentially abundant in SCCOHT tumor samples (blue).
E. (Top) Scatterplot of BIN-67 differential expression data restricted to genes that were preferentially abundant in normal ovarian tissue. (Bottom) Scatterplot of BIN-67 differential expression data restricted to genes that were preferentially abundant in SCCOHT tumor samples. The Wald test followed by BH false discovery rate correction was used to report significance (implemented by the DESeq2 package). The number of genes with |log2FC| > 1 are annotated above each plot.
F. Restoration of SMARCA4 in BIN-67 cells causes gene expression changes that significantly overlap the changes seen between normal ovarian tissue and SCCOHT tumors (One sided Fisher’s exact test, p < 2.2e-16).
Importantly, the gene expression changes upon SMARCA4/2 rescue corroborate transcriptional signatures found in SCCOHT patient tumors. We analyzed tumor RNA-seq from SCCOHT primary tumors from a previous study6 and compared them to transcriptional profiles from normal ovarian tissue29 (Figure 6D). We found that genes that were preferentially abundant in normal ovarian tissue highly overlapped with genes that were upregulated upon the restoration of SMARCA4 or SMARCA2 to BIN-67 cells (Figure 6E), and that genes that were preferentially abundant in SCCOHT tumors were downregulated upon SMARCA4/2 rescue in BIN-67 cells (Figure 6E). This overlap was statistically significant (Figure 6F, one sided Fisher’s exact test, p < 2.2 e-16).
To summarize, by leveraging cell line models of the rare ovarian cancer, SCCOHT, we have uncovered several principles underlying mSWI/SNF complex targeting. In the absence of the SMARCA4/2 subunits, mSWI/SNF complexes are reduced to a residual assembly with a baseline affinity to chromatin but lacking the distinguishing properties of BAF and PBAF subcomplexes. (Supplementary Figure 6). Surprisingly, reintroduction of ATPase-deficient SMARCA4 restores mSWI/SNF localization to ~25% of its target sites and reestablishes differential localization of BAF and PBAF complexes to pre-marked enhancers and transcription initiation sites, respectively. ATPase-driven DNA accessibility is responsible for the remaining 75% of gained sites, particularly for BAF complexes, which also do not require pre-marked H3K4me1 for targeting to these sites. These ATPase activity-mediated events rewire the transcriptional state of SCCOHT cells toward tumor suppressive gene expression programs found in normal ovarian tissue, providing a mechanistic context behind SMARCA4/2 dual loss in SCCOHT.
Methods
Cell lines
BIN-67 cell lines were grown in custom media (20% FBS, 40% DMEM, 40% DMEM/Ham’s F12). SCCOHT-1 cell lines were grown in RPMI supplemented with 10% FBS. SW-13 and HEK-293T cell lines were grown in MEF media (DMEM supplemented with GlutaMax, HEPES, Sodium Pyruvate, and MEF NEAA with 10% FBS). All media was supplemented with pen/strep.
Constructs
The pLEX307 constitutive overexpression backbone was used for all cloning and overexpression experiments. The WT SMARCA4 gene was subcloned from MGC Human SMARCA4 Sequence-Verified cDNA purchased from GE Dharmacon (Accession: BC136644, Clone ID: 9020634). The WT SMARCA2 clone was subcloned from pBABE hBRM, a gift from Robert Kingston (Addgene plasmid # 1961). The K785R SMARCA4 mutant was subcloned from pBJ5 BRG1 DN, a gift from Jerry Crabtree (Addgene plasmid # 17874). The T910M SMARCA4 mutant was created using overlap PCR performed on the WT SMARCA4 clone using a custom set of internal primers to introduce the T910M mutation, which was subcloned into modified pTight (Clonetech) and pLEX307 backbone. Oligonucleotides used for this protocol are included in Supplementary Table 4. A pLEX307 vector expressing GFP was used as an overexpression control.
Lentiviral infection
pspax2 and pMD2.G packaging vectors were co-transfected with pLEX307 vector containing the clone of interest into HEK-293T cells, using PEI as a transfection reagent. Cells were incubated for 72 hours, and the media was filtered with a 0.4 µm filter before being either concentrated with an ultracentrifuge (20,000 RPM for 2.5 hours) or added directly to cells plated at 70% confluence with 1:1000 of 10mg/mL protamine sulfate.
Whole cell lysates
1× 10^6 trypsinized cells were washed twice in PBS and suspended in cold RIPA buffer (10 mM Tris/Cl [pH 7.4], 1 mM EDTA [pH 7.4], 300 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM DTT, 1 mM PMSF) and rotated for 30 minutes at 4°C.
Nuclear extracts
After trypsinization, cells were incubated in Buffer A (25 mM HEPES pH 7.6, 5 mM MgCl2, 25 mM KCl, 0.05 mM EDTA, 10% glycerol and 0.1% NP40 with protease inhibitor (Roche), 1 mM DTT and 1 mM phenylmethylsulfonyl fluoride (PMSF)) for 10 minutes and the pellets were resuspended in 600 μl of Buffer C (10 mM HEPES pH 7.6, 3 mM MgCl2, 100 mM KCl, 0.5 mM EDTA and 10% glycerol with protease inhibitor, 1 mM DTT and 1 mM PMSF) with 67 μl of 3 M (NH4)2SO4 for 20 minutes. The lysates were spun down using a tabletop ultracentrifuge at 100,000 rpm at 4°C for 10 minutes. Nuclear extracts were precipitated with 200 mg of (NH4)2SO4 on ice for 20 minutes and finally purified as pellets by ultracentrifugation at 100,000 rpm at 4°C for 10 minutes. The pellets were resuspended in IP Buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA and 1% Triton-X100 with protease inhibitor, 1 mM DTT and 1 mM PMSF) for the subsequent experiments.
Density sedimentation gradient
Nuclear extract (800 μg, quantified by Bradford) was resuspended in 200 ul of 0% glycerol HEMG buffer (supplemented with protease inhibitors and DTT) and overlaid onto a 11 ml 10%–30% glycerol (in HEMG buffer) gradient prepared in a 14 × 89 mm polyallomer centrifuge tube (331327, Beckman Coulter, Brea, CA, USA). Tubes were centrifuged in an SW40 rotor at 4°C for 16 hr at 40,000 rpm. Fractions (0.550 ml) were collected and used in immunoblot analyses.
Immunoprecipitation
Immunoprecipitations (IP) were performed with 100 ug of nuclear extract resuspended in standard IP buffer (150 mM NaCl, 1 mM EDTA, 1% Triton-X) and rotated overnight with 1.25 ug antibody. Antibodies used were: SMARCA4 (Santa Cruz, G-7; Abcam ab110641 used in Supplementary Figure 2F); SMARCA2, (Bethyl A301–015); SMARCC1 (Santa Cruz, H-76); SMARCD1 (Santa Cruz, 23); ARID2 (Santa Cruz, E-3); PBRM1 (EMD/Millipore, ABE70, Bethyl, A301–591A); ARID1A (Santa Cruz, C1), SS18 (Cell Signaling Technologies, D6I4Z), SMARCC2 (Santa Cruz, E-6), and mouse IgG (Santa Cruz, sc-2025) as a negative control.
Mass spectrometry
For proteomic analysis, SMARCA4, SMARCD1 and SMARCC1 antibody was crosslinked with dimethyl pimelimidate (DMP) to Protein G GammaBind Sepharose beads (GE) before immunoprecipitation from SCCOHT-1 nuclear extract. Bait protein was eluted with high salt and urea buffer, followed by trypsin digest and mass spectrometry (Thermo Exactive Plus Orbitrap).
Purification of complexes
Complexes were purified essentially as previously described with few modifications30. SW-13 and 293T cell lines stably expressing HA-DPF2 constructs were cultured in 150mm dishes and expanded. Cells were scraped and washed with cold PBS. Suspension was centrifuged at 3000 rpm for 5 min at 4°C and pellets were resuspended in hypotonic buffer (HB) containing 10mM Tris HCl pH 7.5, 10mM KCL, 1.5 mM MgCl2, 1mM DTT, 1mM PMSF and incubated on ice for 5min. Nuclear pellets were resuspended in high salt buffer (HSB) containing 50mM Tris HCl pH 7.5, 300mM KCL, 1mM MgCL2, 1mM EDTA, 1mM, 1% NP40, 1mM DTT, 1mM PMSF and protease inhibitor cocktail. Homogenate was incubated on rotator for 1 hour. Homogenates then were centrifuged at 20000 rpm (30000g) for 1 hour at 4°C using SW32Ti rotor. Nuclear extract was filtered through a 0.45um filter and incubated overnight with antibody-bead cocktail prior to elution. Eluted material was then subjected to density gradient centrifugation.
Western blot
Protein was loaded onto Bis-Tris 4–12% gradient Novex gels and run for 150V for 90 minutes. A wet transfer was performed for 2.5 hours at 165 mA at 4 degrees Celsius on to PVDF membrane. After transfer, membranes were blocked in milk for 1 hour at room temperature before applying primary and fluorescent secondary antibodies (Goat Anti-Mouse IgG Antibody, IRDye® 680RD Conjugated, LICOR, Goat Anti-Rabbit IgG Antibody, IRDye® 800CW Conjugated, LICOR) for visualization on a LICOR Odyssey. Antibodies used were: SMARCA4 (Santa Cruz, G-7); SMARCA2, (Bethyl A301–015); SMARCC1 (Santa Cruz, H-76); SMARCC2 (Santa Cruz, E-6); SMARCD1 (Santa Cruz, 23); ARID2 (Santa Cruz, E-3); PBRM1 (EMD/Millipore, ABE70); ARID1A (Santa Cruz, C7), SS18 (Cell Signaling Technologies, D6I4Z), BRD7 (Santa Cruz, B-8), TBP (Abcam, 5184), SMARCB1 (Santa Cruz, A-5), GAPDH (Santa Cruz, G-9), ACTL6A (Santa Cruz, E-3), and CTCF (EMD/Millipore, 07–729).
Immunofluorescence
SW-13 cells expressing SMARCA4 after lentiviral treatment at low multiplicity of infection (MOI) were split to 50% confluency on a 24-well plate. Cells were washed with PBS, incubated in −20 degree C methanol for 5 minutes, then washed twice with IF wash buffer (0.1% NP40, 1 mM sodium azide, PBS 1X) and blocked overnight in blocking buffer (IF wash buffer + 10% FBS). SMARCA4 (Santa Cruz G-7) and SS18 (Cell Signaling Technologies, D6I4Z) antibodies were diluted into blocking buffer and incubated for 3 hours at room temperature. Cells were then washed 3x with IF wash buffer and incubated with secondary antibody for 1 hour (Goat anti-Rabbit IgG Alexa Fluor 546, Cell Signaling Technologies A-11010; Goat anti-Mouse IgG Alexa Fluor Plus 555, Cell Signaling Technologies A32727). Slides were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo, P36935).
Senescence staining
BIN-67 cells treated with SMARCA4 and SMARCA2 lentivirus for two weeks post selection were fixed and stained as described in the Senescence β-Galactosidase Staining Kit (Cell Signaling Technologies) alongside a naïve control.
Generation of knockout HEK-293T cell lines
CRISPR-Cas9 KO constructs targeting SMARCA4 and SMARCA2 were purchased from Santa Cruz and co-transfected into HEK-293T cells using Lipofectamine 3000 reagent. Cells were selected with puromycin at 2ug/ml for 5 days. Single cell clones were selected and screened for loss of subunit expression using immunoblot.
ATP consumption assay
Constructs expressing SMARCA4 wild type, K785R, T910M and an empty vector control were transiently expressed in naïve 293T cells for 72 hours. Cells were trypsinized and pooled in batches of 50 million cells each, followed by PBS wash. Cells were treated with nuclear extract buffer A (25mM HEPES pH8.0, 5mM MgCl2, 25mM KCl, 0.05mM EDTA, 10% glycerol, 0.1% NP40) and resuspended in 200–300 uL of lysis buffer (50mM Tris pH 8.0, 150mM NaCl, 0.1% NP40) with protease inhibitor and incubated in cold room with constant rotation for 30 minutes. Cell debris was discarded via ultracentrifuge at 100K RPM for 5 minutes. Supernatant was collected, quantified and used for subsequent overnight immunoprecipitation step with 30uL of V5 bead slurry in cold room with constant rotation.
On the following day, IPs were washed three times with 1ml of wash buffer (10mM Tris pH 7.5, 50mM NaCl, 5mM MgCl2) and resuspended in 25uL of complete 1X ATPase Assay Reaction Buffer (10mM Tris pH 7.5, 50mM NaCl, 5mM MgCl2, 20% glycerol) with reaction reagents (0.1mg/ml BSA, 1mM DTT, 4mM ATP, 01–0.5ug/ul DNA) and transferred to PCR tubes. Consuption reaction was incubated at 37C for 1 hour. Following ATP hydrolysis, beads were separated from assay, and 20uL of ATPase reaction was transferred to an opaque white 384-well plate. 20uL of room temp ADP-Glo reagent was added and incubated at room temp for 40 minutes. Then, 40uL of ADP-Glo detection substrate was added to reaction and incubated for 1 hour. Luminescence was recorded on SpectraMax with an integration time of 0.25–1 second/well.
Differential salt extraction
Differential salt extraction was performed as previously described21.
Genetic interactions (MINGLE)
To call SMARCA4/2 dual loss genetic interactions from screening data, we utilized the MINGLE framework16 with a few modifications. CRISPR screening data and cell line data were downloaded from the Depmap Public 18Q2 dataset (https://depmap.org/portal/download/all/). Cell lines were called as SMARCA4/2 dual deficient from literature references (BIN-67, COV434, OVK18)17 or if the cell line exhibited low gene expression of SMARCA2 (RPKM < 2) and exhibited biallelic damaging mutations on SMARCA4, and did not exhibit a strong dependency on either gene (CERES score > −0.5) (H1581, H23). Differential dependency was calculated directly from CERES scores, and significance was assessed using a Wilcox rank-sum test, and p-values were adjusted for multiple comparisons with the Benjamini-Hochberg correction. Differential dependencies with FDR < 0.25 were reported as significant. Pi scores and graphical output were performed as in the MINGLE pipeline.
ChIP-seq
mSWI/SNF chromatin immunoprecipitation was performed using a modified version of protocol that was previously described21,31,32. In brief, cells were fixed in 1% formaldehyde at 37°C for 10 minutes, quenched in 2.5 M glycine, and snap frozen. Cells were thawed and lysed and the prepared nuclei was placed in a Covaris miliTube and sonicated for 20 minutes on a Covaris AFA Sonicator at standard settings (140 PIP, 5 W, 10% duty factor). The equivalent of 6–10 million cells were used in each immunoprecipitation. Three micrograms of the following antibodies were used: SMARCA4 (Abcam ab110641); DPF2 (Abcam, ab134942); SMARCC1 (homemade rabbit antibody raised against amino acids 998–1073 of human protein); SS18 (Cell Signaling Technologies D6I4Z); ARID2 (Cell Signaling Technologies D8D8U); H3K4me1 (Abcam, ab176877); H3K27ac (Abcam, ab4729); H3K27me3 (Cell Signaling Technologies 9733S); H3K9me3 (Abcam, ab8898), MLL3/4 (homemade33). IP’s were washed in 150 and 500 mM NaCl wash buffers as well as a LiCl wash. Protein-DNA fragments were eluted using an SDS/DTT buffer and reverse crosslinked overnight. DNA was captured with SPRI beads (Agilent). Rubicon library prep was used to generate libraries, and libraries were sequenced on Illumina NextSeq 500. ChIP-seq immunoprecipitations for each antibody were performed in singlicate across each of the rescue conditions; overlap of antibodies targeting two distinct epitopes or subunits were used for analyses.
ChIP analysis
For alignment of ChIP-Seq data, Bowtie2, version 2.1.0 was used to map reads to the hg19 human reference genome, using the parameter –k 1. Raw reads were filtered for redundant reads and aligned to the hg19 genome. All downstream analysis was performed on bam files with duplicates removed using the samtools rmdup command.
Peaks were called using MACS 2.0 at a q value cutoff of 0.001 using the narrow peak setting, with the exception of ARID2 peaks, which were called using the broad peak setting. Peaks that fell in ENCODE blacklisted regions or were mapped to unmappable chromosomes (not chr1–22, X or Y) were removed. ChIP-seq track densities were generated per million mapped reads with MACS2 using the –B –SPMR options and visualized using IGV. Overlaps for ChIP venn diagrams were created using the ChIPPeakAnno bioconductor package.
ChIP peaksets
All peaksets used in the paper were derived from at least two complementary ChIP-seq experiments (using antibodies designed to two distinct epitopes (subunits) on mSWI/SNF complexes) from the same experimental condition.
The residual complex peakset was defined as the union between peaks identified in the BIN-67 + control rescue (any of ARID2, DPF2 or SMARCC1 peaks). Both control DPF2 and ARID2 peaks analyzed in Figure 2 were intersected with SMARCC1 peaks from the same experimental condition. Both SS18 and PBAF peaks upon rescue of SMARCA4 in Figure 2 were intersected with SMARCA4 peaks from the same experimental condition.
BAF activity-independent peaks were defined as SS18 peaks (intersected with SMARCA4 peaks as noted above) that were also found in the SS18 ChIP performed in SMARCA4 K785R rescue. The remaining SS18 peaks were annotated as activity-dependent. PBAF activity-independent peaks were similarly defined but with ARID2 instead of SS18. Enhancers (Figure 4A) were defined as H3K4me1 peaks in the WT SMARCA4 rescue condition that overlap ATAC-seq peaks in the same condition.
ChIP occupancy scatterplots
Read count across peak sets of interest were calculated by calling the Rsubread bioconductor package function featureCounts() on duplicate removed bam files. These values were divided by the total number of mapped reads divided by one million, giving a normalized value of reads per million mapped reads for each interval in the input bed. Scatterplots were plotted with the R function smoothScatter.
ChIP feature distributions
Peaksets were analyzed with the ChIPSeeker package using default settings34.
ChIP heatmaps
Heatmaps of ChIP occupancy were generated created using HTSeq. To account for the 200bp average fragment length selected for in sonication, fragment length was extended 200 bp from the edge of each genomic interval. Total read counts for each interval were normalized to reads per million mapped reads (RPM). For each antibody the resulting matrix has a width of 4 kb and a height of the number of peaks in the indicated set. Strandedness of the interval was not considered except for the direction TSS plots (Figure 4D, Supplementary Figure 4A). Heatmaps were visualized using matplotlib in python. Heatmaps were ordered by the maximum value in each matrix row of the indicated antibody.
ATAC-seq
ATAC-seq was performed as described previously35. In particular, 50,000 cells were harvested for each condition as suggested. Libraries were prepared using the suggested barcodes and sequenced on a paired end Illumina NextSeq 500, with 4 samples on a single paired end lane. Sequencing data was aligned as previously described36. All rescue and control experiments were performed in duplicate, and sequencing data merged into a single file for processing.
RNA-seq
One million cells of each condition were harvested using Qiagen RNeasy kit and 1 ug was DNase treated and used in Illumina TruSeq Stranded mRNA library prep kit. All samples were validated to have RIN > 0.6 on BioAnalyzer. Libraries were sequenced on Illumina NextSeq 500. All rescue conditions were performed in replicate, with the SMARCA4 rescue performed in triplicate.
After sequencing, RNA-seq data reads were mapped using default parameters to hg19 using STAR version 2.5.2a. Differential genes were called using DESeq2. RPKM values were calculated using GFOLD version 1.1.4. Unless otherwise noted, log2 fold change and Bonferri-corrected p values were generated using DESEQ2, with reads mapped using RSUBREAD. Genes were considered significantly changing if they had an adjusted pvalue <.05 and a log2 fold change of at least 2.
Clustering of differentially expressed genes
The log2 fold change of differentially expressed genes in the SMARCA4 rescue condition were plotted with the pheatmap R package across all four experimental conditions tested (SMARCA4 K785R, SMARCA4 T910M, SMARCA4 WT and SMARCA2 WT). Clustering was performed with complete linkage clustering and Euclidian distance, and a cluster number of 3 was chosen for further analysis.
GO enrichment
GO term enrichment was performed with the GOrilla web tool (http://cbl-gorilla.cs.technion.ac.il)37, using each of the three differentially expressed gene clusters as target gene lists, and the set of expressed genes among BIN-67 RNA-seq experiments (RPKM > 2, resulting in 13,189 genes) as background.
Target gene assignment and enrichment
Genes were annotated as targets of BAF regulation if there was at least one BAF peak within 30 kb of the TSS of that gene. Genes were annotated as PBAF targets in the same fashion. Target genes were split between activity-dependent and activity-independent groups based on which peakset was more abundant within the 30 kb window around the TSS.
For each of the three differentially regulated gene clusters, overrepresentation of BAF and PBAF target genes was assessed with a one-sided Fisher’s exact test against the background of all expressed genes (13,189 genes total).
GSEA and enrichment of target genes
Geneset enrichment analysis was performed with the GSEA software38 on the MSigDB C2 genesets using differential expression data upon SMARCA4 WT reintroduction. Genesets that were enriched for upregulated genes with a FWER p-val < 0.01 were used for downstream analysis. Overrepresentation of BAF activity-dependent or BAF activity-independent target genes among upregulated genes within these genesets was calculated with a one-sided Fisher’s exact test against the background of all upregulated genes (533 total).
Primary tumor and normal ovarian data
Raw RNA abundance counts for 10 primary SCCOHT tumors (SRP052896) and 108 primary ovary samples (GTEx) were computed using the recount2 pipeline39. [PMID: 28398307] Due to considerable batch differences between the two samples, we used a more conservative non-parametric rank-based method for identifying differentially expressed genes. Specifically, the top and bottom 5% of differentially ranked genes between conditions were identified for further analysis.
Supplementary Material
Acknowledgements
We thank members of the Kadoch Laboratory for helpful conceptual and experimental advice throughout the development of this study. We thank the DFCI Molecular Biology Core Facility, particularly Z. Herbert, for library preparation and sequencing, G. Boulay for advice regarding ChIP-seq optimization, and the Taplin Mass Spectrometry Facility for mass spectrometry analysis and data processing. We thank B Vanderhyden (Ottawa Hospital Research Institute) and Ralf Hass (Hannover Medical School) for the provision of the BIN-67 and SCCOHT-1 cell lines, respectively. This work was supported in part by funding from the National Science Foundation Graduate Research Fellowship (2015185722) and NIH T32 Training Grant in Genetics and Genomics to J.P, the NIH DP2 New Innovator Award 1DP2CA195762-01(C.K.), the American Cancer Society Research Scholar Award RSG-14-051-01-DMC (C.K.), the Pew- Stewart Scholars in Cancer Research Grant (C.K.).
Footnotes
Competing Interest Statement
C.K. is a Scientific Founder, fiduciary Board of Directors member, Scientific Advisory Board member, consultant, and shareholder of Foghorn Therapeutics, Inc., (Cambridge, MA).
Data Availability Statement
All sequencing data is available at GEO: GSE117735. Code to generate figures is available at https://github.com/joshbiology/sccoht. All peaks called from raw data can be found at https://figshare.com/s/d74eccb73f20af21a6da. All custom-defined peaksets can be found at https://figshare.com/s/00ac067cf47edea9805d.
Statistics and Reproducibility
The following panels are representative of experiments repeated the following number of times:
Figure 1A – duplicate
Figure 1C – DPF2, 10+ replicates, SW-13, triplicate
Figure 2B – duplicate of both conditions
Supplementary Figure S1A – 10+ replicates
Supplementary Figure S1B – duplicate
Supplementary Figure S1C – duplicate
Supplementary Figure S1D – singlicate
Supplementary Figure S1E – SW-13 triplicate, DPF2 10+ replicates, SMARCD1 10+ replicates
Supplementary Figure S2A – singlicate
Supplementary Figure S2B – singlicate
Supplementary Figure S2D – duplicate (second shown in Supplementary Figure 1F)
Supplementary Figure S2E – duplicate
Supplementary Figure S2F – duplicate with two diff. antibodies (both shown on same blot)
Supplementary Figure S2G – duplicate
Supplementary Figure S3A – 5+ replicates
Supplementary Figure S3B – singlicate
Supplementary Figure S3D – singlicate
Supplementary Figure S5C – representative of 3 images from same experiment
Additional information can be found in the Life Sciences Reporting Summary.
References
- 1.Kadoch C & Crabtree GR Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science advances 1(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnezis AN et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. The Journal of pathology 238, 389–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jelinic P et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet 46, 424–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos P et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet 46, 427–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witkowski L et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet 46, 438–43 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Le Loarer F et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nature genetics 47, 1200–1205 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Karnezis AN et al. Loss of switch/sucrose non-fermenting complex protein expression is associated with dedifferentiation in endometrial carcinomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 29, 302–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Upchurch KS, Parker LM, Scully RE & Krane SM Differential cyclic AMP responses to calcitonin among human ovarian carcinoma cell lines: A calcitonin-responsive line derived from a rare tumor type. Journal of Bone and Mineral Research 1, 299–304 (1986). [DOI] [PubMed] [Google Scholar]
- 9.Gamwell LF et al. Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics. Orphanet journal of rare diseases 8, 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otte A et al. A tumor-derived population (SCCOHT-1) as cellular model for a small cell ovarian carcinoma of the hypercalcemic type. Int J Oncol 41, 765–75 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Leibovitz A, McCombs WM, Johnston D, McCoy CE & Stinson JC New human cancer cell culture lines. I. SW-13, small-cell carcinoma of the adrenal cortex. Journal of the National Cancer Institute 51, 691–697 (1973). [PubMed] [Google Scholar]
- 12.Mashtalir N et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 175, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson BG et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Molecular and cellular biology 34, 1136–1144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman GR et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences of the United States of America 111, 3128–3133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyers RM et al. Computational correction of copy-number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature genetics (2017). [DOI] [PMC free article] [PubMed]
- 16.Rauscher B et al. Toward an integrated map of genetic interactions in cancer cells. Molecular Systems Biology 14, e7656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan-Penebre E et al. Selective Killing of SMARCA2- and SMARCA4-deficient Small Cell Carcinoma of the Ovary, Hypercalcemic Type Cells by Inhibition of EZH2: In Vitro and In Vivo Preclinical Models. Molecular cancer therapeutics 16, 850–860 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Wang Y et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. The Journal of pathology 242, 371–383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middeljans E et al. SS18 Together with Animal-Specific Factors Defines Human BAF-Type SWI/SNF Complexes. PLoS ONE 7(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadoch C et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45, 592–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama RT et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nature genetics 49, 1613–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raab JR, Resnick S & Magnuson T Genome-Wide Transcriptional Regulation Mediated by Biochemically Distinct SWI/SNF Complexes. PLOS Genetics 11(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodges HC et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nature structural & molecular biology 25, 61–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dykhuizen EC et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bultman SJ, Gebuhr TC & Magnuson T A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes & development 19, 2849–2861 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Local A et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nature Genetics 50, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Levine SS, Boyer LA, Jaenisch R & Young RA A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorighi KM et al. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Molecular cell 66, 568–5760000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium G Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, N.Y.) 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 30.Mashtalir N et al. Autodeubiquitination Protects the Tumor Suppressor BAP1 from Cytoplasmic Sequestration Mediated by the Atypical Ubiquitin Ligase UBE2O. Molecular Cell 54, 392–406 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Takaku M et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome biology 17, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride MJ et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell (2018). [DOI] [PMC free article] [PubMed]
- 33.Cao K et al. An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Science advances 4(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G, Wang L-G & He Q-Y ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology 109, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denny SK et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 166, 328–342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collado-Torres L et al. Reproducible RNA-seq analysis using recount2. Nature Biotechnology 35, 319–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.