Figure 1.
Biochemical and genomic characterization of a residual complex in SMARCA4/SMARCA2 dual-deficient cancer cell lines.
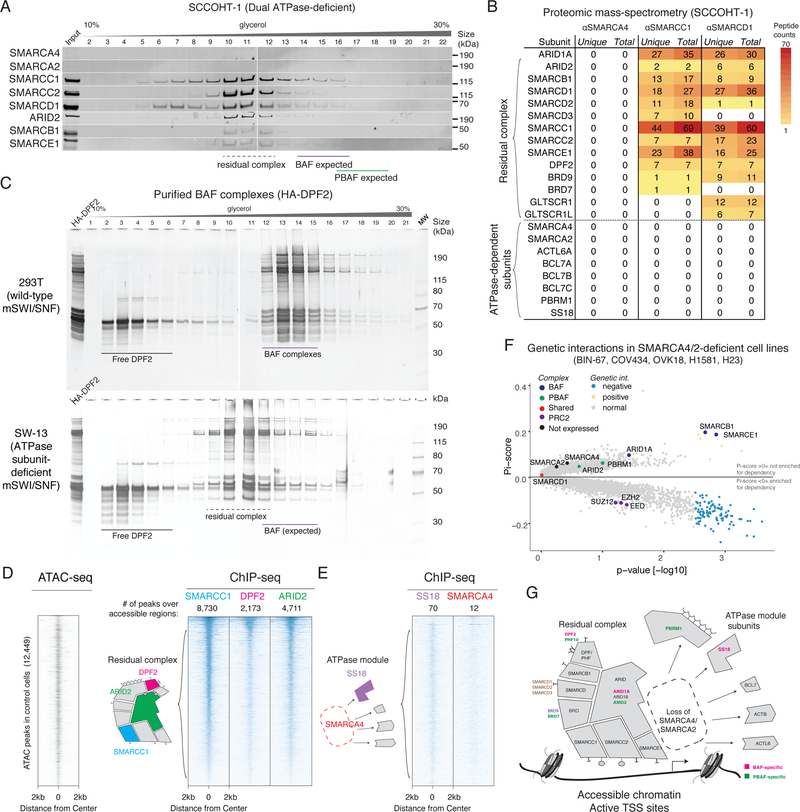
A. Density sedimentation and immunoblot performed on SCCOHT-1 nuclear extracts. The residual mSWI/SNF complexes of both BAF and PBAF types exhibit similar elution profiles. See also Supplementary Figure 7a.
B. Peptides corresponding to mSWI/SNF subunits identified in immunoprecipitation/mass-spectrometry from SCCOHT-1 nuclear extract. SMARCC1 and SMARCD1 immunoprecipitations identify a cohort of subunits which remain stably bound to complexes in the absence of both ATPase subunits.
C. Density sedimentation performed on endogenously-purified BAF complexes from SMARCA4/2-intact (293T) and SMARCA4/2-deficient (SW-13) cell lines, using an HA-tagged DPF2 subunit as bait.
D. Heatmaps of chromatin accessibility (ATAC-seq) and residual mSWI/SNF occupancy (ChIP-seq) in the BIN-67 cell line, treated with a control Luciferase vector. Each row of the heatmap corresponds to a peak from the union of ATAC-seq and SMARCC1 ChIP-seq peaks in this condition. Rows are rank ordered by SMARCC1 occupancy. Each column shows the normalized read density for different experimental conditions across a 4-kb window centered at each peak.
E. Heatmaps reflecting SS18 and SMARCA4 ChIP-seq experiments performed in the BIN-67 cell line.
F. Genetic interaction data derived from CRISPR-Cas9-based screens performed in SMARCA4/2-dual-deficient and SMARCA4/A2-intact cell lines. Significance was calculated by a two-sided Wilcox test on the SMARCA4/2 deficient lines (n=5) against the WT lines (n=386). An FDR cutoff was 0.25 after BH correction was used.
G. Schematic of residual mSWI/SNF complex composition in the absence of the SMARCA4/2 ATPase subunits.