Figure 3.
Rescue of a catalytic activity-deficient ATPase module is sufficient to localize PBAF and BAF complexes to a subset of target sites genome-wide.
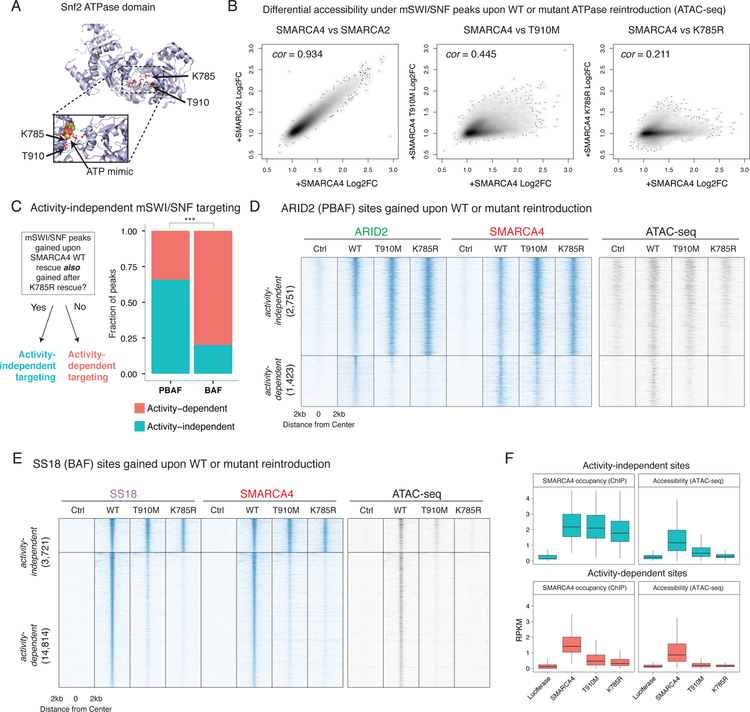
A. Mapping of T910M and K785R mutant residues to the ATP binding pocket of the yeast Swi2/Snf2 crystal structure (as performed in Hodges et al., 2018, NSMB) shows a proximity to a crystalized ATP mimic. Top: Overall structure of the Swi2 domain, mutated residues highlighted. Bottom: Enhanced view of ATP binding pocket in the presence of an ATP mimic.
B. Scatterplots of differential accessibility under SMARCC1 peaks upon rescue of different ATPase alleles. Each point in the scatterplot corresponds to a SMARCC1 peak called in the BIN-67 SMARCA4 rescue condition. Each axis displays the fold change in accessibility under each peak (as measured by ATAC-seq reads per million mapped to that peak) as compared to the Luciferase condition. Spearman correlation is shown.
C. Stacked bar plot of the proportion of BAF and PBAF peaks that are activity-dependent and activity-independent. Fisher’s two-sided exact test performed on total ChIP-seq peaks (n = 22,709), *** p < 1e−3
D. Heatmaps of chromatin accessibility and mSWI/SNF occupancy across ARID2 peaks gained upon rescue of a SMARCA4 allelic series. The ARID2 peakset is split into two groups: activity-independent peaks, and activity-dependent peaks. Rows are rank ordered by ARID2 occupancy in the wild type rescue condition.
E. Heatmaps of chromatin accessibility and mSWI/SNF occupancy across SS18 peaks that are gained upon rescue of a SMARCA4 allelic series. The SS18 peakset is split between activity-independent peaks, and activity-dependent peaks. Rows are rank ordered by SS18 occupancy in the wild type rescue condition.
F. Boxplots showing normalized readcounts across SS18 activity-independent (n=3,721) and activity-dependent peaks (n=14,814). First and third quartiles are denoted by lower and upper hinges. Whiskers extend to the smallest and largest values within 1.5x the interquartile range from the lower and upper hinges.