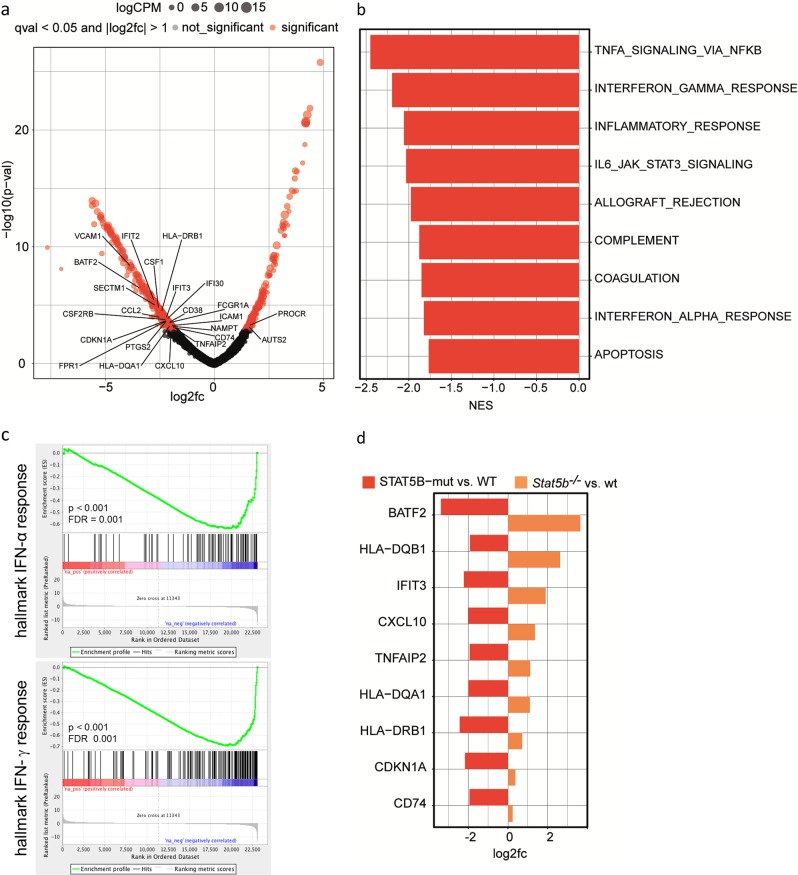
Fig. 7.
IFN responses are enhanced in human STAT5B mutant T cell large granular lymphocyte leukemia (T-LGLL). a RNA-seq from STAT5B mutant (n = 4) and wt (n = 13) T-LGLL patient samples. Significantly differentially (q value <0.05 and fold change >2) upregulated (n = 256) or downregulated (n = 376) genes were identified. Deregulated genes belonging to IFN-α and IFN-γ hallmark gene sets are indicated. Size of points corresponds to logCPM (counts per million) expression levels. b Significantly (NES <−1.75, FDR <0.25) negatively enriched hallmark gene sets obtained from GSEA of log 2 fold-change ranked gene lists from differential expression analysis of STAT5B mutant (vs. wt) T-LGLL cases. c Enrichment plots of the IFN-α and IFN-γ hallmark gene sets from GSEA. d Genes significantly (p-adjust <0.05) upregulated in Stat5b−/− BCR/ABLp185 cell lines and significantly (p-adjust <0.05) downregulated in STAT5B mutant T-LGLL cases. IFN interferon, STATB signal transducers and activators of transcription 5B, RNA-seq RNA-sequencing, wt wild type, GSEA geneset enrichment analysis, NES normalized enrichment score, FDR false discovery rate