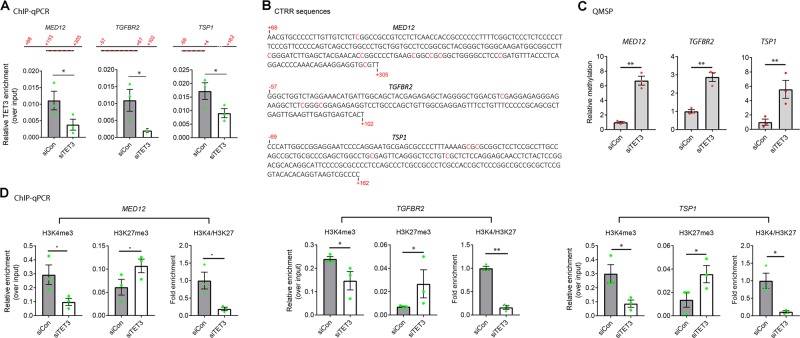
Fig. 3.
TET3 affects DNA methylation and histone modifications of the MED12, TGFBR2, and TSP1 promoters. a UtLM cells were transfected with siCon or siTET3 for 48 h, followed by ChIP-qPCR analysis. Data are presented as mean relative TET3 enrichment over input. n = 3. Red numbers indicate nucleotide positions relative to the transcriptional start sites, with PCR products depicted as red-stripped bars. b Sequences of critical transcription regulatory regions (CTRR) of MED12, TGFBR2, and TSP1. The differentially methylated cytosine residues are marked in red. The red numbers mark the positions of the indicated nucleotides relative to the transcriptional start sites. c UtLM cells were transfected with siCon or siTET3 for 48 h, followed by QMSP analysis. n = 3. d UtLM cells were transfected with siCon or siTET3 for 48 h, followed by ChIP-qPCR analysis. Data are presented as mean relative enrichment over input. n = 3. All data are representative of at least two independent experiments and are presented as mean ± SEM. *p < 0.05, **p < 0.01