Fig. 5.
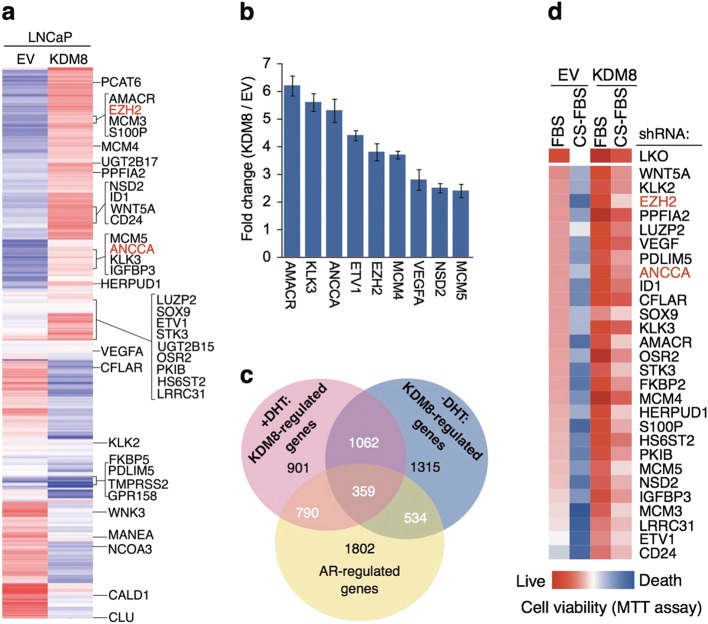
KDM8 controls specific subsets of androgen-responsive program. a Heat map of hierarchical clustering of androgen-regulated genes (ARGs). LNCaP-overexpressed KDM8 and EV control cells were cultured in the presence or absence of 1 nM DHT followed by gene expression profiling with microarray analysis. The ARGs that differentially expressed by KDM8 (≥1.5-fold) and the positions for well-known ARGs are indicated. Two genes EZH2 and ANCCA selected for ChIP assay are marked in red. b qRT-PCR analysis of the selected ARDs. LNCaP cells (KDM8 and EV) were cultured in the androgen-deprived media for 3 days followed by qRT-PCR analysis. Triplicate experimental data were expressed as fold change as compared to EV control after normalizing to internal control 16S rRNA. c Venn diagram showing the overlap of KDM8-regulated genes and ARDs. d Heat map showing cell viability of LNCaP cells (KDM8 and EV) by knocking down the indicated ARGs in the absence of androgen. LNCaP cells infected with the shRNAs targeting the ARGs as indicated were then cultured in media containing normal FBS or charcoal-stripped FBS (CS-FBS) for 3 days. Cell viability was assessed by MTT assay. EZH2 and ANCCA selected for chromatin immunoprecipitation (ChIP) assay are marked in red