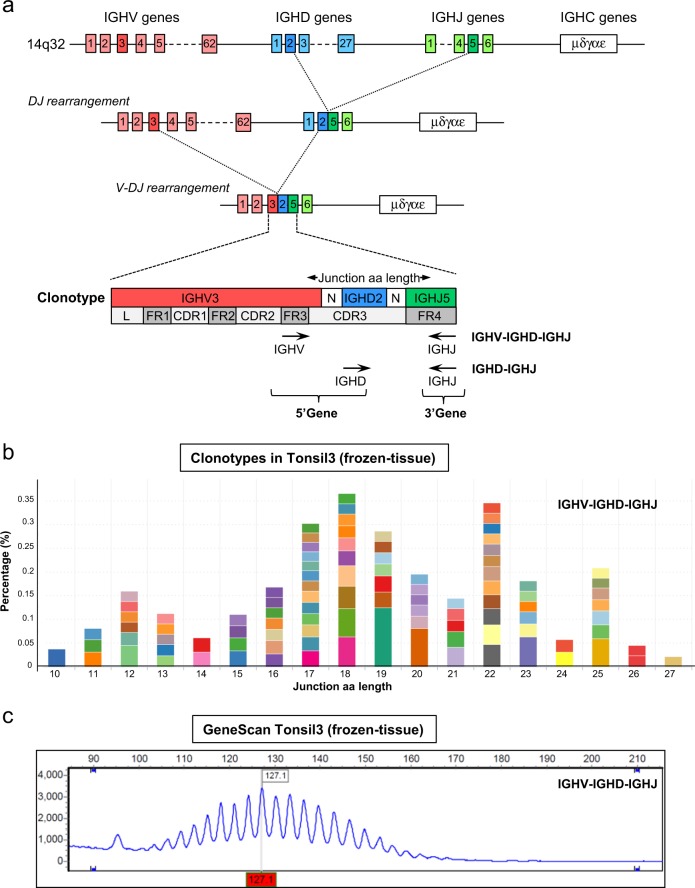
Fig. 1.
Identification of distinct clonotypes by next generation sequencing (NGS)-based detection of immunoglobulin heavy chain rearrangements. a Schematic representation of the immunoglobulin heavy chain (IGH) locus on chromosome 14q32, indicating the presence of 62 IGHV genes, 27 IGHD genes, 6 IGHJ genes, and 5 IGHC genes. Following RAG-mediated DJ and V-DJ rearrangement, each B cell generates one (productive rearrangement) or two (a non-productive and productive rearrangement) specific clonotypes, each consisting of one IGHV, IGHD, and IGHJ gene segment. For the detection of IGHV-IGHD-IGHJ gene rearrangements the forward IGHV primers located in framework region 3 (VH FR3) are combined with IGHJ reverse primers. For the detection of IGHD-IGHJ rearrangements the forward IGHD and reverse IGHJ primers will generate amplicons. Successful amplification will yield DNA fragments that cover the junctional region with a specific amino acid (aa) length, representing the CDR3 region and anchor points. The leader region (L), the conserved framework regions 1–4 (FR1–4) and the hypervariable complementary regions 1–3 (CDR1–3) are indicated. b Detection of distinct clonotypes in DNA isolated from a frozen tonsil tissue of a healthy donor (Tonsil3), indicating a polyclonal pattern of IGHV-IGHD-IGHJ rearrangements with variable junction aa length. Each colored bar indicates a distinct clonotype, and the 100 most abundant clonotypes are shown. The NGS data were analyzed and visualized by ARResT/Interrogate. c GeneScan profile of Tonsil3 IGHV-IGHD-IGHJ rearrangements as determined with EuroClonality/BIOMED-2 PCR