Figure 3. Common phenotypes associated with histone and non-histone substrates of KMT activity.
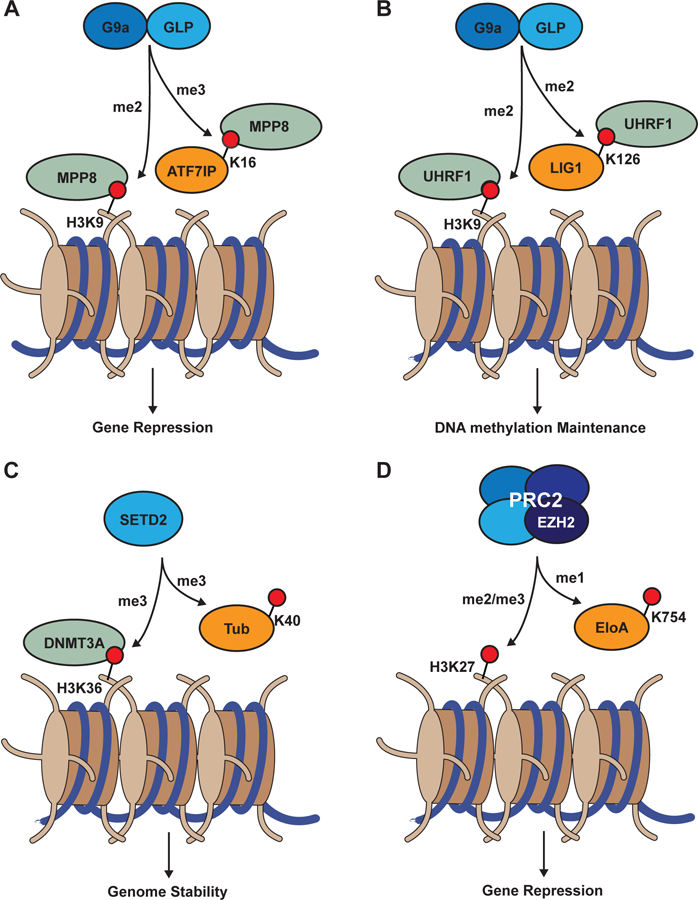
(A) MPP8 chromodomain recognition of G9a/GLP-dependent H3K9 and ATF7IPK16 methylation is associated with gene silencing. (B) UHRF1 tandem Tudor domain recognition of G9a/GLP-dependent H3K9 and LIG1 methylation is associated with DNA methylation inheritance. (C) SETD2-dependent H3K36 and αTubulinK40 methylation contributes to genomic stability. (D) The catalytic subunit of the PRC2 complex, EZH2, methylates H3K27 and ElonginAK754 to promote gene silencing.
