Figure 4. Technologies developed to reveal KMT substrates.
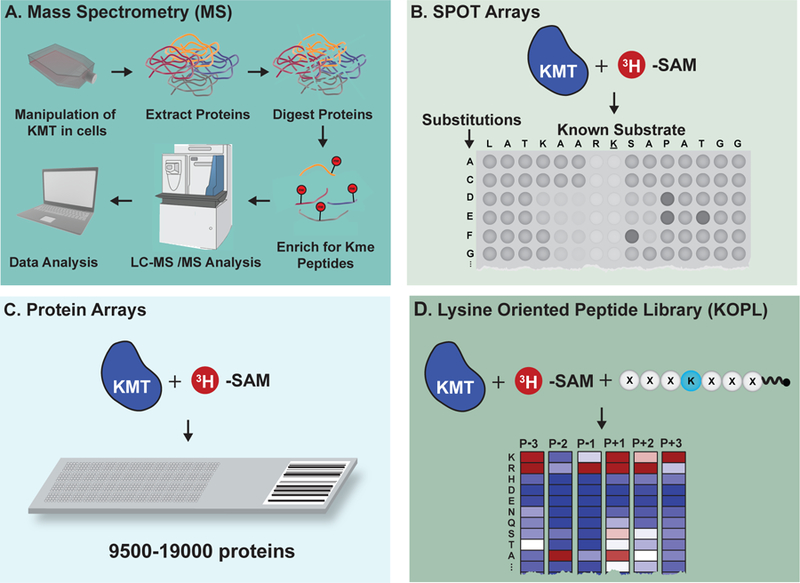
(A) Typical mass spectrometry (MS)-based proteomics pipeline for the identification of lysine methylation sites from cultured cells. (B) SPOT arrays evaluate the sequence tolerance of known KMT peptide substrates. (C) Protein arrays facilitate de novo identification of KMT substrates. (D) Lysine-oriented peptide libraries (K-OPL) reveal the sequence determinants of KMT substrate selectivity.
