Figure 5. Perturbation of Transcriptional Landscape Alters the Chromosomal Distribution of ClsN and Chromosome Domain Organization.
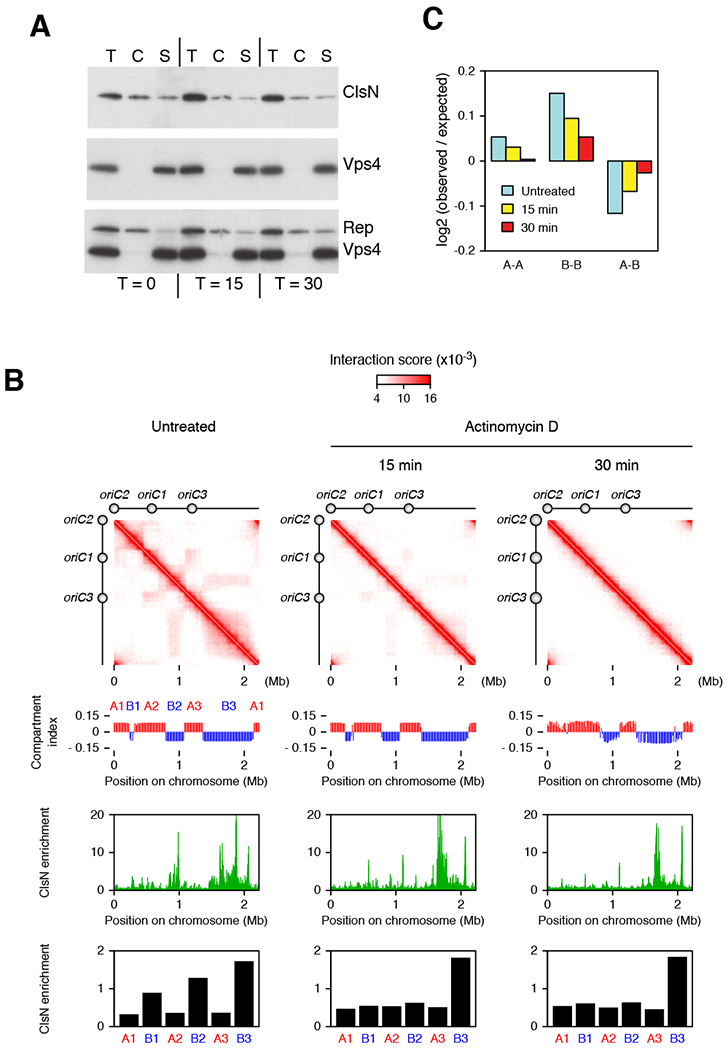
(A) Chromatin fractionation of ClsN in S. acidocaldarius after treatment with actinomycin D. After separating cell extract into chromatin (C) and soluble (S) fractions, total protein (T) and extract fractions were run on SDS-PAGE. ClsN and soluble and chromatin markers, Vps4 and Reptin respectively, were detected by western blotting. Protein samples were collected before actinomycin D treatment and 15 and 30 minutes after treatment.
(B) Changes in chromosome conformation and ClsN distribution in S. acidocaldarius across a time course of actinomycin D treatment. Top panels: Hi-C contact matrices. Upper middle panels: compartment index plots, the A compartment is in red, the B compartment is in blue. Lower middle panels: ClsN ChIP-seq profiles at 1-kb resolution. All samples were normalized to the input DNA from the corresponding time point. Bottom panels: Enrichment of ClsN in the domains divided by the in the domains. ClsN enrichment is calculated as ChIP read coverage normalized to input for the domains. The location of each domain is based on the untreated condition.
(C) Distance-normalized frequency at the various timepoints of genomic contacts within the A compartment (A-A), within the B compartment (B-B) and between A and B compartments (A-B), based on the compartment identities in the untreated sample. See also Figure S7.
