Figure 7. Locus Specific Effects on ClsN Occupancy Upon Constitutive Expression of a Regulon.
Two biological replicates per sample were used for analyses.
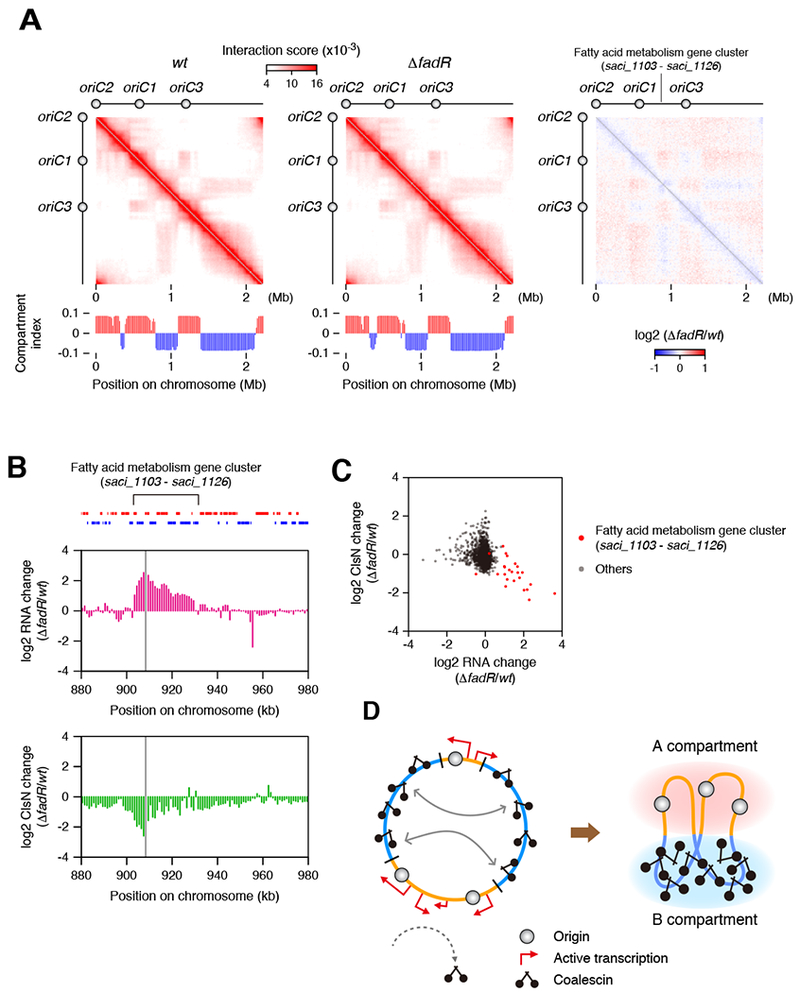
(A) Hi-C analysis in the wild-type and ΔfadR deletion strains of S. acidocaldarius.
(B) RNA-seq and ClsN ChIP-seq profiles in the vicinity of the fatty acid metabolism regulon. Log2-fold changes in RNA level (RPKM) and ClsN enrichment in ΔfadR were quantified for each 1-kb bin. The bin containing the fadR gene was omitted from the plots and shaded in gray. Locations of genes are shown on top (gene orientations are indicated by different colors).
(C) Log2-fold changes in RNA level (TPM) and ClsN enrichment in ΔfadR were quantified for protein-coding genes and shown as a scatter plot.
(D) Model for the organization of Sulfolobus chromosomes. Transcription excludes ClsN from the origin-containing A-domains. ClsN binding to the B-domains promotes coalescence of these regions. Coalescence could be promoted either by protein-protein interactions or via a loop-extrusion/catenation activity.
