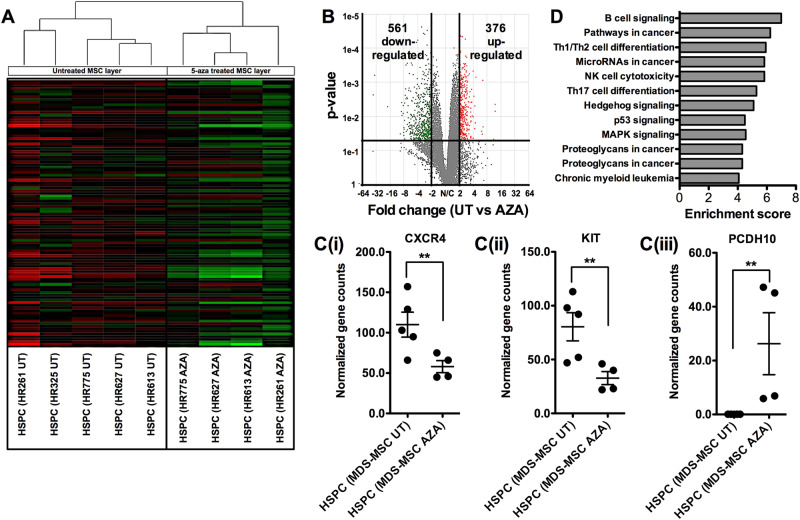
Fig. 4.
RNA-seq analysis of healthy CD34+ HSPCs after co-culture with different MDS-MSCs. a Hierarchical clustering of HSPCs co-cultured with MDS-MSCs (HR261, HR325, HR613, HR627 and HR775). HDS-MSCs are treated with AZA (AZA) or untreated (UT). HSPCs from HR325 AZA co-cultures were insufficient for this analysis. Clustering of differentially expressed genes show distinct groupings between HSPCs that were co-cultured on UT vs AZA MDS-MSCs. HSPCs co-cultured with AZA treated HR261 MDS-MSCs were less similar to HSPCs from other treated systems, similar to previous observations (Fig. 3). b There are more down-regulated genes in HSPCs from UT vs AZA MDS-MSCs. Using a p < 0.05 and FC > 2, 561 vs 376 genes were down- vs up-regulated, respectively. c Normalized gene counts from gene specific analysis (Partek Flow) of CXCR4 (i), KIT (ii) and PCDH10 (iii). CXCR4 and KIT are commonly seen up-regulated in leukemic blasts and PCDH10 is a tumor suppressor that is silenced by hypermethylation. d KEGG pathway analysis of differentially expressed genes in HSPCs after co-culture with UT vs. AZA MDS-MSCs indicates activation of cancer/aberrant differentiation signaling networks in healthy HSPCs after exposure to dysplastic MDS-MSCs. *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant. Non-paired student’s t test was performed. All data represented as mean ± SEM