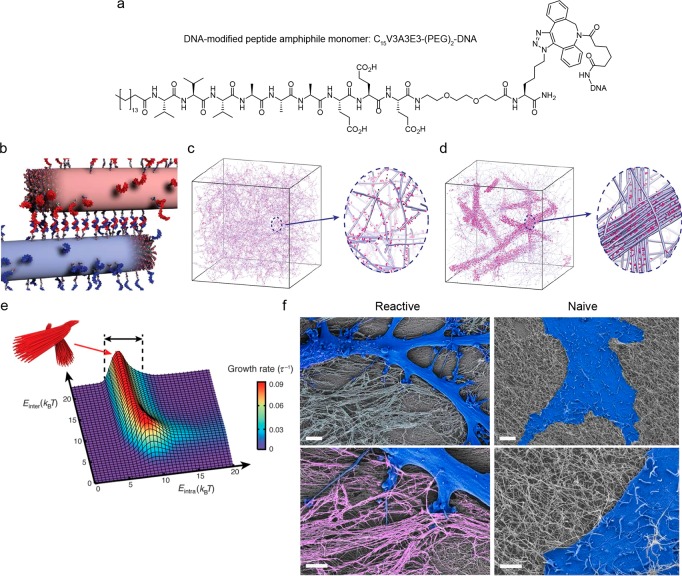
Figure 4.
Reversible formation of superstructures in peptide amphiphile–DNA networks. (a) Structure of a peptide amphiphile (PA) monomer modified with oligonucleotides via DIBAC conjugation. (b) PA copolymers of inert and DNA-modified monomers cross-link due to hybridization of complementary grafted oligonucleotides which subsequently cluster within the fibers. (c) Simulations show that homogeneous hydrogels are obtained when molecular exchange of DNA monomers between PA fibers is prohibited. The magnified view shows individual fibers (blue) with a stochastic distribution of DNA monomers (pink) along the fibers. (d) When molecular exchange is allowed, the simulations result in the formation of bundled fibers. The magnified view shows a bundle of fibers (blue) enriched with DNA (pink) in a matrix of individual fibers depleted of DNA monomers. (e) Simulations showing that the bundle growth rate is a function of the molecular attraction between monomers and the interfiber hybridization of the grafted oligonucleotides (respectively, the intra- and interfiber energies (Eintra, Einter)). Bundles form within the energy range 5 kBT < Eintra. (f) SEM micrographs of astrocytes cultured on cross-linked, bundled (left), and individual (right) PA-DNA fibers. The cells display a reactive phenotype on the bundled fibers while a naive phenotype is observed on the individual fibers. Cells are falsely colored in blue and bundled PA-DNA fibers in pink. The magnified view (lower images) shows the cell–substrate interaction. Scale bars represent 5 μm (upper images) and 2 μm (lower images). Adapted and reproduced with permission from ref (31). Copyright 2018, American Association for the Advancement of Science.