Figure 7.
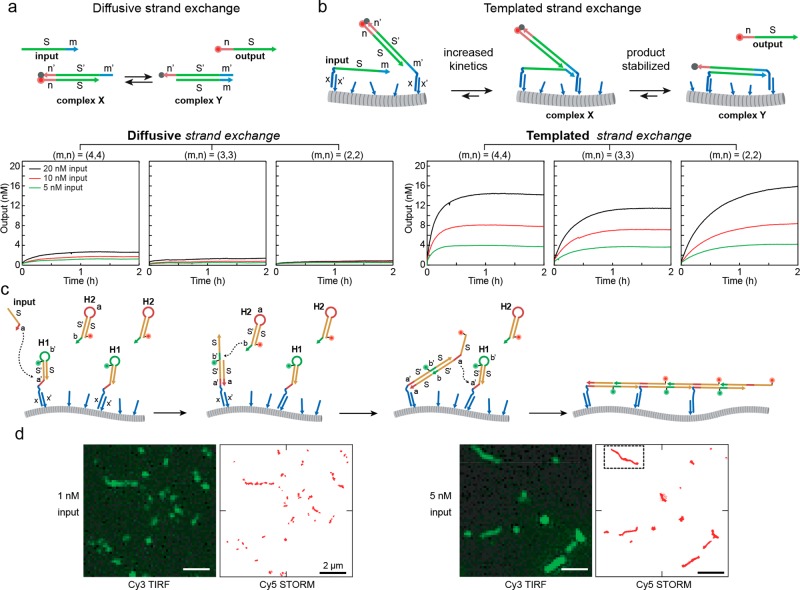
Acceleration of DNA-based computing on supramolecular polymers. (a) Schematic representation of a strand exchange reaction using freely diffusing oligonucleotide reactants with the kinetic characterization of a reaction with toeholds of various lengths. (b) Schematic representation of a strand exchange reaction templated by a BTA polymer. The reactants are recruited to the polymer via the hybridization of a sequence complementary to the oligonucleotide on the BTA. This results in a high local concentrations which consequently increases the association kinetics of toehold binding. Additionally, due to the multivalent anchoring of the product to the supramolecular polymer, the product is stabilized resulting in increased operation yields. (c) Schematic representation of a hybridization chain reaction (HCR) templated by the BTA polymers. Starting upon the addition of an input, the sequential opening of two labeled metastable hairpin strands results in the formation of a polymeric DNA assembly. (d) TIRF images of immobilized BTA polymers containing Cy3 labeled monomers with corresponding STORM images of the HCR product containing Cy5 labeled oligonucleotides after the HCR was initiated by addition of, respectively, 1 and 5 nM input strand. The scale bars represent 2 μm. Adapted and reproduced with permission from ref (35).