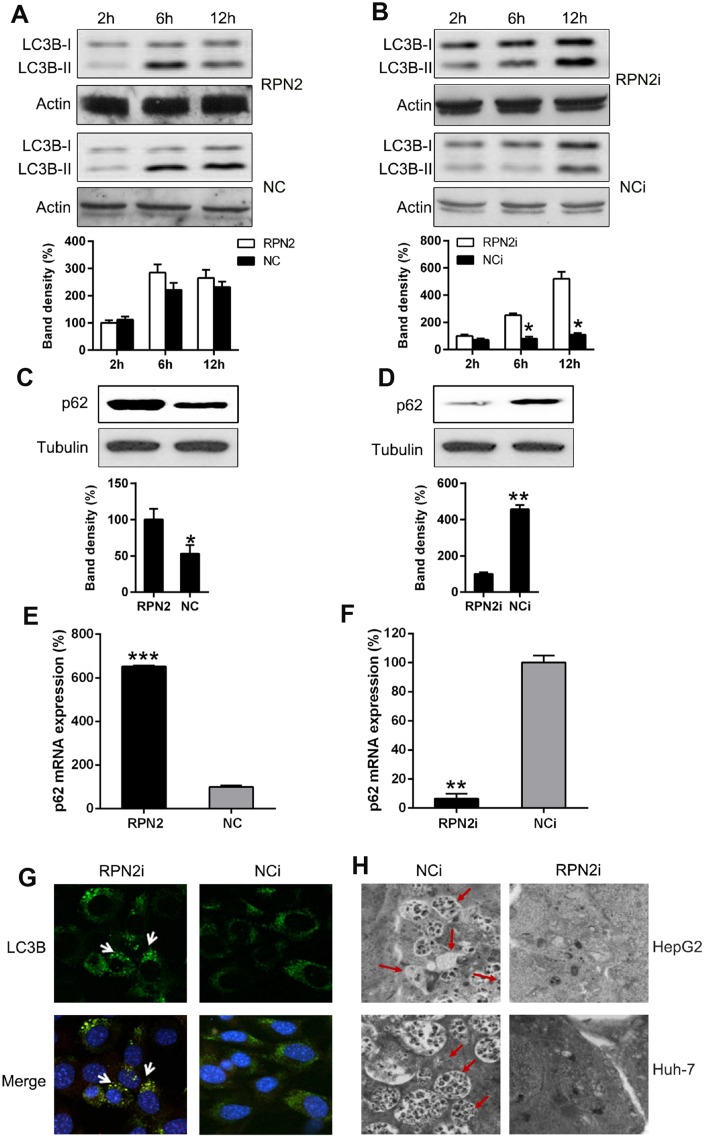
Figure 6.
RPN2 depletion leads to autophagy. Cells were firstly serum-starved for 24 hr. Then cells were treated with the solvent (DMSO) or 10 nM 3-Methyladenine (3-MA) for the indicated time before harvesting. (A, B) HepG2 cells were seeded onto 12-well plates, kept overnight and then starved for one day. Cells were infected with AD-RPN2 or transfected with shRNA-RPN2, and were serum-starved for 24 hr. Then cells were treated with the solvent (DMSO) or 10 nM 3-Methyladenine (3-MA) for the indicated time before harvesting. Whole cellular proteins were subjected to WB to indicate dynamic of LC3B Western blot analysis then determined the expression level of (C, D) p62 in cells with RPN2 overexpression and RPN2 silencing. (E, F) qPCR assay analyzed mRNA expression of p62 during RNP2 overexpression and silencing, respectively. (G) HepG2 cells were co-transfected with the shRNAs and GFP-LC3, as indicated, for 2 days. Cellular location of GFP-LC3B was then observed by IFA (magnification: 400x). (H) Autophagesomes were shown by transmission electron microscopy. Electron microscopy images of autophagic vacuoles in HepG2 and Huh-7 cells with silenced RPN2 or controls. Red arrows illustrate some of the autophagic vacuoles at different stages of the autophagy process (magnification: 2000x). The band of target protein was normalized to the density of action. The quantification was performed independently in a single band. The experiments were performed three times. Data were recorded as mean ± SD. **P <0.01, ***P <0.001 vs control group.