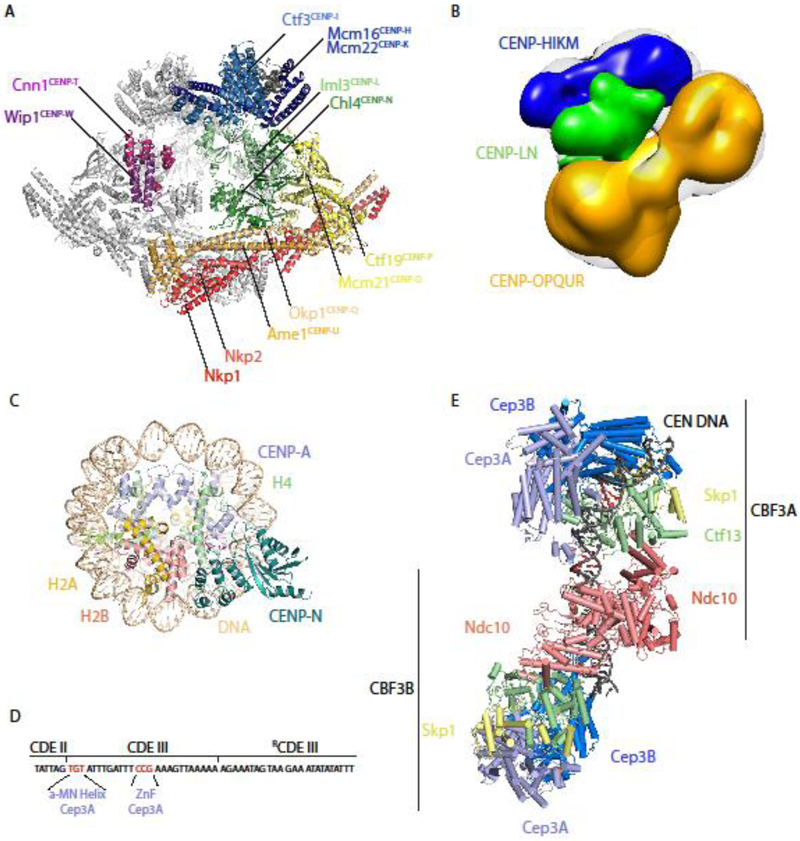
Figure 3. Recently published structures of inner kinetochore protein complexes.
(A) Dimers of the 13-protein S. cerevisiae Ctf19c flank a central cavity. CENP-NLCh14/Im13 occupies the middle of each protomer, and its CENP-ACse4-binding domain extends into the central cavity (PDB ID: 6NUW) [29]. (B) An ~22 Å negative stain 3D reconstruction of a reconstituted 11-protein human CCAN, in which CENP-HIKM (density of subcomplex in blue) and CENP-OPQUR (density of sub-complex in yellow) sandwich CENP-NL (density of subcomplex in green) [32]. (C) The N-terminus of human CENP-N specifically recognizes centromeric, CENP-A-containing nucleosomes by binding to the unique L1 loop (indicated in red) of CENP-A [27, 28]. (Model shown from PDB ID: 6C0W, related structures: 6BUZ and 6EQT) (D) Schematic of the 56 bp nuclease-resistant sequence bound to CBF3. Conserved TGT and CCG motifs in Centromere Determining Element III (CDEIII) are specifically bound by the α-MN helix and zinc finger (ZnF) motifs, respectively, of the same Cep3A subunit [32]. (E) Budding yeast CBF3 binds centromeric DNA as a dimer, recognizing specific DNA sequences (colored red) prior to Cse4 deposition [32]. Orange spheres indicate zinc atoms. (Model shown from PDB ID: 6GYS, similar structures: 6GSA and 6F07).