Fig. 3.
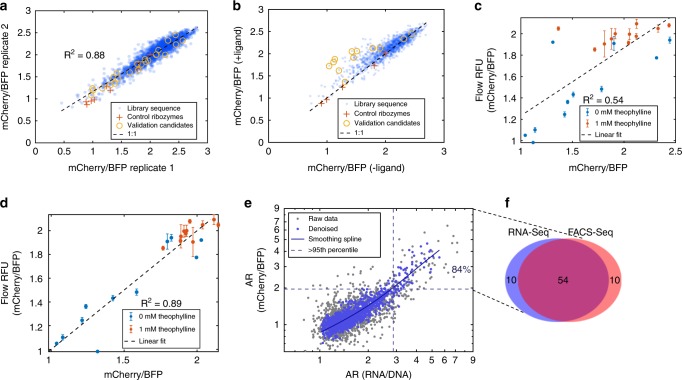
FACS-seq assay of a theophylline ribozyme switch library. a Relative fluorescence values from replicate FACS-Seq experiments (R2 = 0.87) on a log10 scale. Each dot represents a unique sequence with over 100 sequencing read counts in an experiment (1202 sequences). Pluses are spiked-in control ribozymes; circles indicate sequences selected for validation. b Relative fluorescence values from a FACS-Seq assay performed on a theophylline ribozyme switch library in the absence and presence of ligand. −/+ Ligand conditions are 0/1 mM theophylline. c Relative fluorescence values from flow cytometry analysis of individual sequences plotted against relative fluorescence values from a FACS-seq assay (R2 = 0.55). Error bars indicate standard deviation of two biological replicates. d Relative fluorescence values from flow cytometry analysis of individual sequences plotted against denoised relative fluorescence values from a FACS-seq assay (R2 = 0.89). Error bars as in c. e Activation ratios of relative fluorescence values from FACS-seq plotted against activation ratios of normalized RNA read count from RNA-Seq. Raw data were denoised using a regression model from AutoML; from the denoised data both assays agree on ~84% of sequences with >95th percentile activation ratio. Dashed lines indicate the 95th percentile AR thresholds. f Venn diagram shows the number of sequences shared between RNA-Seq and FACS-Seq for sequences with >95th percentile activation ratio. a, c, d All R2 are based on log10 transformed values. Source data are available in the Source Data file