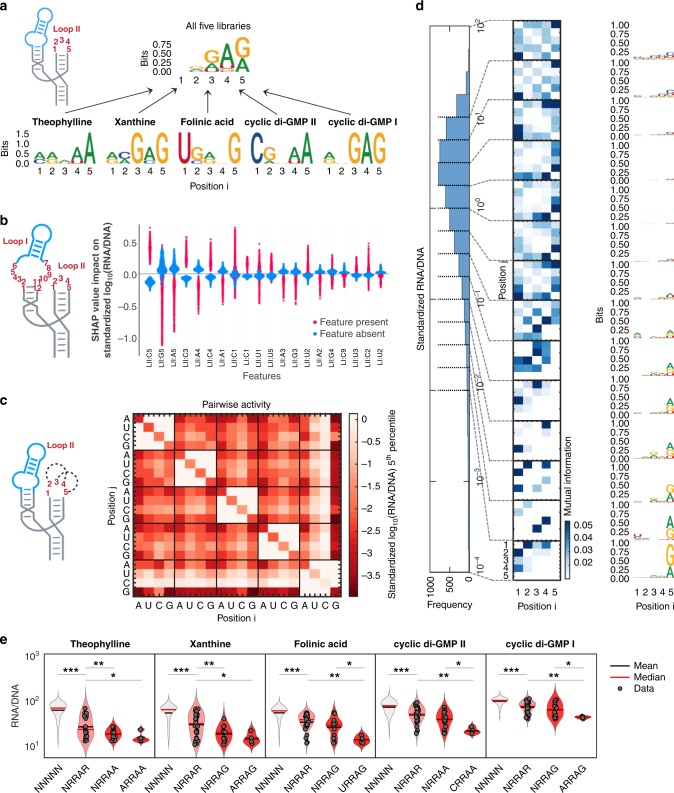
Fig. 5.
Sequence-function relationships and design principles. a Sequence logos representing Shannon entropy and consensus for five sequences with the lowest RNA/DNA levels from each loop II N5 library and for the combination of all five libraries. Schematic illustrates the position numbering on loop II. b SHAP (SHapley Additive exPlanation) values indicating the impact of the 20 most important sequence features (identity and position) on the predicted standardized log10(RNA/DNA) value of every sequence (dot) from five pooled libraries (n = 5093 N5 library variants), derived from a boosting tree regression model (XGBoost). Sequence features are derived from loop I and loop II as indicated with position numbering in the adjacent schematic. c Pairwise contribution to the 5th percentile standardized log10(RNA/DNA) ratio of the five pooled libraries for each of A, U, C, G nucleotides at two positions within loop II. Schematic indicates position numbering of the feature analyzed, with dashed lines as examples of pairwise interactions. d The five pooled libraries divided into 14 activity bins according to different levels of standardized log10(RNA/DNA). For sequences in each bin, mutual information analysis is indicated for a pair of nucleotides in loop II in the heat map, and the sequence logo of the sequences in the same bin is shown to the right. e Distribution of normalized mRNA levels RNA/DNA of different ligand libraries when constrained by conserved loop II N5 motifs indicated in the x-axis. N indicates the presence of all bases A, U, C, G; R indicates the presence of only A and G; asterisks indicate significance from one-tailed t-tests, *p < 0.05, **p < 0.005, ***p < 10−9. Filled circles are individual sequence data points