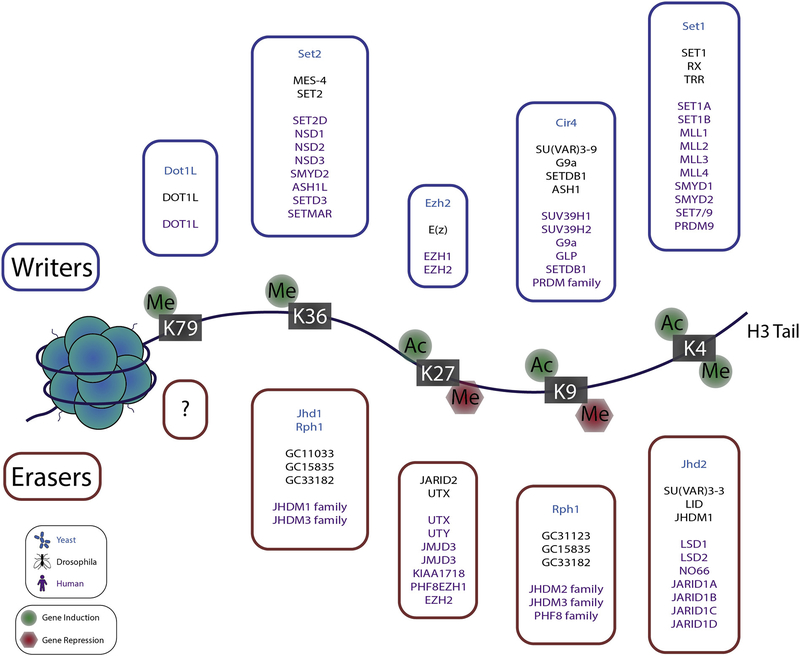
Figure 1. Illustration of histone H3 lysine modification effect on gene expression.
The nucleosome is composed of double stranded DNA wrapped around a “hairy hockey puck” comprised of core histone proteins and their unstructured N- and C- terminal tails (H3 tail is exaggerated for illustration purposes). Acetylation and methylation of lysine residues on core histone proteins are mutually exclusive modifications on the same residue of the same subunit. Histone lysine acetylation is associated with increased gene expression at the indicated residues. Histone lysine methylation is associated with gene activation when found at H3K79, H3K36 and H3K4 but is generally associated with gene silencing at other residues. In the boxes, a list of writers and erases is included.