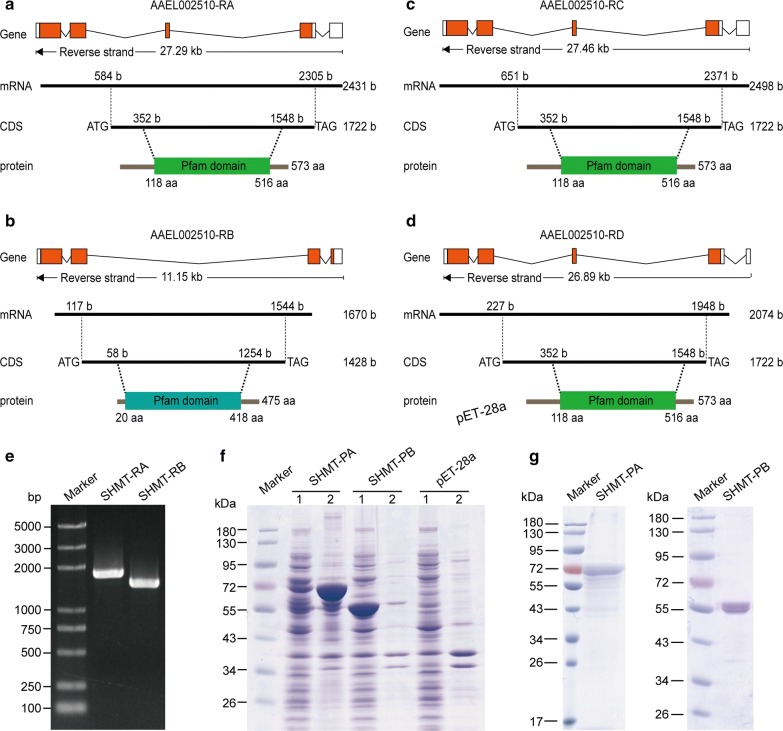
Fig. 1.
Gene structures, isoforms and prokaryotic expression patterns of A. aeSHMT. All sequences were downloaded from VectorBase at https://www.vectorbase.org/organisms/aedes-aegypti. a–d Schematic diagrams of isoforms A, B, C and D. The square box, black line and red box in the gene structure above represent the exon (mRNA), intron and protein coding region (CDS), respectively. These four SHMT isoforms correspond to four different lengths of mRNA and two different lengths of CDS or protein. The Pfam domain was generated by online software Smart at http://smart.embl-heidelberg.de/smart. e Cloning of the CDS for SHMT-RA and SHMT-RB. Two different lengths of CDS are shown in the 1% agarose gel as PCR products of SHMT-RA and SHMT-RB. f SDS-PAGE showing the SHMT proteins produced by prokaryotic expression. Lanes 1: detection of the supernatant; Lanes 2: detection of the precipitate. g SDS-PAGE showing the purified proteins used for the preparation of primary antibody. SHMT-PA was purified only through nickel affinity chromatography, while SHMT-PB was purified through nickel affinity chromatography and by AKTA Purifier 100 (GE Healthcare) (see “Methods”)