Fig. 11.
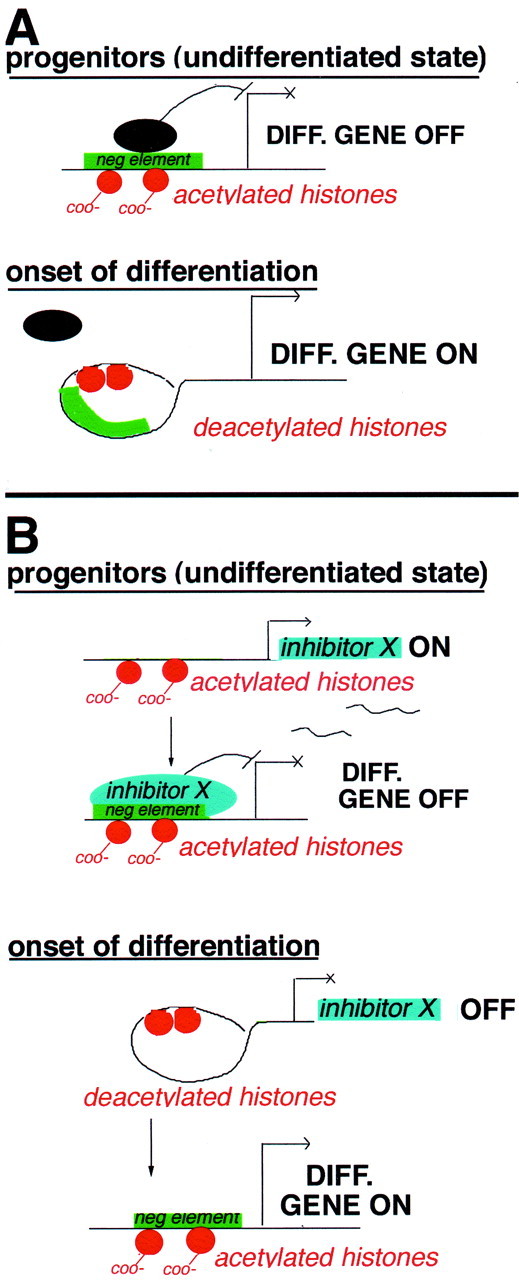
A proposed model of the role of histone deacetylation in oligodendrocyte differentiation. A, Direct model. The first model assumes that histone deacetylation occurs directly in the promoter of specific differentiation genes (e.g., myelin genes). In the presence of mitogens, nucleosomal histones are acetylated on lysine residues (orange circles with COO− tails), and this results in open chromatin conformation and exposure of negative regulatory sites (green rectangles) in the promoter region of myelin genes. Binding of specific regulatory molecules (black ovals) to these sites is favored, and progenitors are kept in an undifferentiated state. On recruitment of HDAC, induced by the withdrawal of mitogens, histone tails are deacetylated (orange circles with no tails), and the chromatin around the negative regulatory sites is compacted, thus preventing the access to transcriptional inhibitors. This event results in onset of differentiation caused by compaction of negativecis-element on the promoter region of myelin genes.B, Indirect model. The second model predicts that histone deacetylation occurs on the promoter of genes encoding for differentiation inhibitors (e.g., gene X). In progenitor cells, nucleosomal histones in the promoter region of the differentiation inhib-itor are acetylated (orange circles with COO− tails), resulting in open chromatin conformation and expression of the inhibitor X. This in turn may bind to negative regulatory sites on differentiation genes and prevent their expression. Onset of differentiation in this case is initiated by recruitment of HDAC to the promoter region, resulting in chromatin compaction and decreased expression of the differentiation inhibitor. According to this model, the negative element on the promoter of differentiation genes may be acetylated but inactive, because of the absence of gene X (left), or deacetylated and compacted (right). In both cases, the expression of differentiation genes is activated by histone deacetylation.
