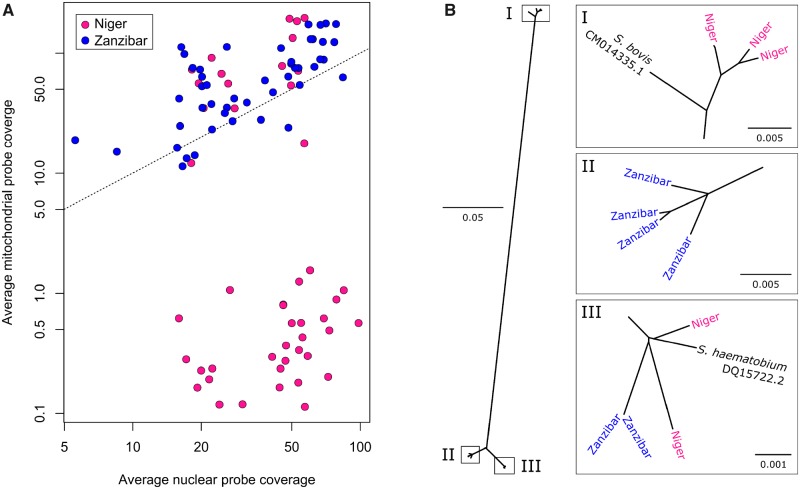
Fig. 2.
Mitochondrial sampling and phylogeny. (A) The mitochondrial genome was sequenced to high coverage despite large variations in the number of reads generated for most samples. For 31 Nigerien samples, mitochondrial reads were at very low frequency regardless of sequencing depth. This was attributed to the inefficient capture of the highly diverged mitochondria in these 31 Nigerien samples. (B) Maximum likelihood phylogeny of 12 mitochondrial genomes from Schistosoma haematobium and S. bovis and haematobium reference sequences. Three Nigerien S. haematobium samples are related to S. bovis indicating mitochondrial introgression from S. bovis into the Nigerien S. haematobium population.