Supplemental Digital Content is available in the text
Keywords: Human gastrointestinal tract, Microbial biodiversity and composition, Beneficial and harmful microorganisms, Metabolite concentrations
Abstract
Background:
Emerging evidences have indicated that the composition of gut microbiota was significantly influenced by central nervous system diseases. The digestion and metabolism disturbances of patients with amyotrophic lateral sclerosis (ALS) might be strongly associated with ALS; however, this has rarely been evaluated in these populations. This study was to evaluate bacterial and archaeal composition of gut flora and the corresponding metabolism performance of these micro-organisms in fecal samples of patients with ALS.
Methods:
A comparative study was performed on the intestinal microbiota from eight patients with ALS and eight healthy individuals at Huadong Hospital during November 2017 to April 2018; meanwhile, the metabolite concentrations of human endotoxin, short-chain fatty acids (SCFA), NO2-N/NO3-N, and γ-aminobutyric acid were also evaluated by spectrophotometry methods. The correlations between intestinal microbiota and metabolite concentration were compared between the two groups using one-way analysis of variance; the relative abundance of beneficial and harmful micro-organisms in fecal samples was also analyzed.
Results:
In general, the richness and evenness of bacterial and archaeal communities of healthy individuals were healthier than that of patients with ALS. The phylum Firmicutes/Bacteroidetes ratio, genus Methanobrevibacter showed an enhancive tendency in patients with ALS, whereas the relative abundance of beneficial micro-organisms (genera Faecalibacterium and Bacteroides) presented a significant decrease tendency in patients with ALS. In addition, the average concentrations of human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid in patients with ALS and healthy individuals were 64.2 vs. 65.3 EU/mL, 57.5 vs. 55.3 μg/mL, 5.7 vs. 5.3 ng/mL, and 6.1 vs. 5.4 μmol/L, respectively, indicating that the digestion and metabolism functions of gastrointestinal tract of patients might decline with this disease.
Conclusions:
The relative abundance of beneficial and harmful micro-organisms respectively showed decrease and increase tendency in patients with ALS.
Introduction
Amyotrophic lateral sclerosis (ALS), which results in a loss of neurons at all levels of the motor system, has been considered as a fatal neuromuscular disease.[1,2] It has been investigated that the crude annual incidence rate of ALS was 2.16 per 100,000 person-year in European.[3] Owing to the ALS is an age-dependent disease, as the population grows and ages, the incidence population might show an enhancive trend around the world.[4] The clinical and scientific interest to study on this disease started to raise in about 1990s, the discovery of survival in ALS was mainly dependent on rate of disease progression, clinical presentation (phenotype), early presence of respiratory failure, and nutritional status of patients.[5,6] Unfortunately, there is no powerful intervention that can be used to change the course of this disease.[7] The count showed that approximately 50% of the patients could survive only below than 3 years, while only about 20% of patients could survive for 5 years and a small percentage could be alive after 10 years.[8] Hence, developing further understanding about disease progression and pathogenesis mechanism of ALS is of great significance and urgency.
The human gastrointestinal tract provides an optimal environmental condition for the growth and metabolism of bacterial and archaeal communities. It was generally accepted that the biodiversity and composition of these micro-organisms were significantly related to human metabolism, immunity and gut-brain axis diseases.[9–11] In literature, the variation of microbial biodiversity and communities showed apparent interactions with the disease progression of central nervous system.[12] In particular, the central nervous system could modify the gastrointestinal tract via the release of hormones, immune and neurotransmitters factors.[13,14] Meanwhile, the level of metabolites (short-chain fatty acids [SCFA], NO2-N/NO3-N, and γ-aminobutyric acid) generated by gut bacteria could positively and negatively influence on autism spectrum disorders.[15] It was observed that the abundance of methanogens showed a significant increase in patients with irritable bowel syndrome diseases.[16,17] However, there are large gaps on the interaction mechanism of gut flora composition and metabolism performance in gastrointestinal tract of patients with ALS.
Owing to ALS is a systemic wasting disease, the patient's weight declined with the digestive disturbances and intestinal barrier dysfunction.[18] It was generally accepted that the gut flora composition and metabolism performance were significantly related to the digestive mechanism of intestines.[19] It has been indicated that the relative abundance of harmful bacteria (genus Dorea) and beneficial micro-organisms (genus Anaerostipes, Oscillibacter, and Lachnospiraceae), respectively presented an enhancive and declined trend in the gastrointestinal tract of patients with ALS.[2] In addition, the host-bacterial interactions were apparently reduced in ALS mice treated with butyrate.[4] The SCFA could be used as the metabolic substrates by Methanobrevibacter genus to produce methane (CH4), which resulted in the weight loss of patients with intestinal diseases.[20] However, there has not been a comprehensive understanding of the interactions about bacterial and archaeal communities and metabolites composition in the human gastrointestinal tract of patients with ALS.
The objective of this study was to evaluate bacterial and archaeal composition of gut flora and the corresponding metabolism performance of these micro-organisms in fecal samples of patients with ALS. The composition of bacteria and archaea, and the concentration of human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid were determined. We hypothesized that the biodiversity and composition of bacteria and archaea and the corresponding metabolism concentration in human gastrointestinal tract might be affected by this disease.
Methods
Ethical approval
The study on microbial community and metabolites composition of patients with ALS was approved by the Ethical Committee of Huadong Hospital of Fudan University (Shanghai) (No. 2017K058). All participants have been told research details before signed informed consent, and all experiments were done in compliance with the approved guidelines and national directives.
Patient's recruitment
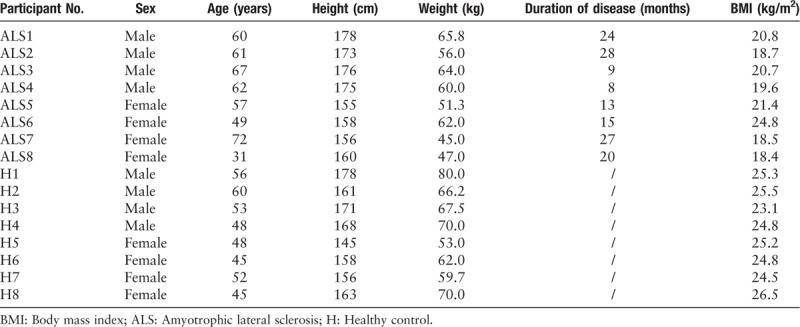
Eight patients with ALS were recruited according to the revised E1 Escorial criteria (1998)[21] at Huadong Hospital from November 2017 to April 2018. The inclusion criteria could be concluded as follows: (1) the survival period of all patients with ALS was more than 3 months; and (2) no antibiotics were used in the last 1 month. The exclusion criteria could be concluded as follows: (1) patients with ALS were with serious heart, lung, liver, kidney, brain and tumor, blood system diseases, and other disorders of life signs; (2) other diseases (ie, cervical spondylosis, spinal cord tumors) that need to be differentiated from ALS; and (3) patients with overeating and drinking in the past 2 weeks. In addition, eight healthy individuals were recruited as the controls at about the same time. The sex, height, weight, duration of disease, and body mass index information of patients with ALS and healthy controls were collected [Table 1].
Table 1.
Sex, age, height, weight, duration of disease, and BMI information of patients with ALS and healthy individuals.
Fecal sampling and polymerase chain reaction amplification
The fecal sample of each participant was collected from the first fecal motion of 1 day after a 30-day similar diet. Then all samples were stored at –80°C. For polymerase chain reaction (PCR) amplification of bacterial communities, the universal bacterial primers of 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) were used to amplify the V4–V5 hypervariable regions, while for PCR amplification of archaeal communities, the universal bacterial primers of 524F_10_ext (5′-TGYCAGCCGCCGCGGTAA-3′) and Arch958R_mod (5′-YCCGGCGTTGAVTCCAATT-3′) were used. The pre-treatment, amplification processes, pyrosequencing data, and quality control of microbial samples were performed as described in previous study.[22] All PCR amplification of microbial samples were performed on the Illumina Miseq PE300 platform (Shanghai Majorbio Bio-Pharm Technology Co. Ltd., China). The archaeal throughput sequencing in fecal sample of patients ALS6 and ALS8 was failed because of sequencing technology reasons.
Metabolites determination
The correlation coefficients (R2) of calibration curves of human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid were determined; absorptivity values on 450 nm wavelength with spectrophotometry, and correlation coefficients (R2) were more than 0.99. The concentrations of human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid of fecal sample were respectively tested with the corresponding enzyme-linked immunosorbent assay (ELISA) kits according to standard directions. Each sample was determined with a two to four repetitions.
Data and statistical analyses
To ensure the high-quality of sequence reads, all reads were checked and denoised based on the listed criteria: (1) set the window options of 50 base pairs (bps), if the average mass value in the window was less than 20, the back end bps was removed from the window, and reads below 50 bps after quality control was filtered to remove reads containing N base; (2) according to the relationship between overlap and paired-end reads, pair reads were merged into a sequence, the minimum overlap length was 10 bps; (3) the maximum mismatch rate of the overlap region of the merged sequence was 0.2; (4) the samples were identified according to the barcode and primers at the end of the sequence. The mismatch number of barcodes was 0 and the maximum primer mismatch number was 2. Then, a similarity of 97% was used to classify the similar 16S rRNA reads into same operational taxonomic units (OTUs) with uclust algorithm.[23] The relative abundance of phylum, class, order, family, and genus was generated according with OUTs annotation, and then determined with a proportion of all reads for each microbial sample. In addition, the biodiversity estimators (Ace, Chao, Shannon, and Simpson) were used to evaluate richness and evenness of microbial communities.[2,22]
In this study, the statistical difference of metabolites concentrations (human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid) between patients with ALS and healthy individuals was compared using one-way analysis of variance (least significant difference test). The data were analyzed using software SPSS version 24.0 (IBM Corp., Armonk, NY, USA). A P < 0.05 was set as the significant level.
Results
Microbial biodiversity and reads coverage
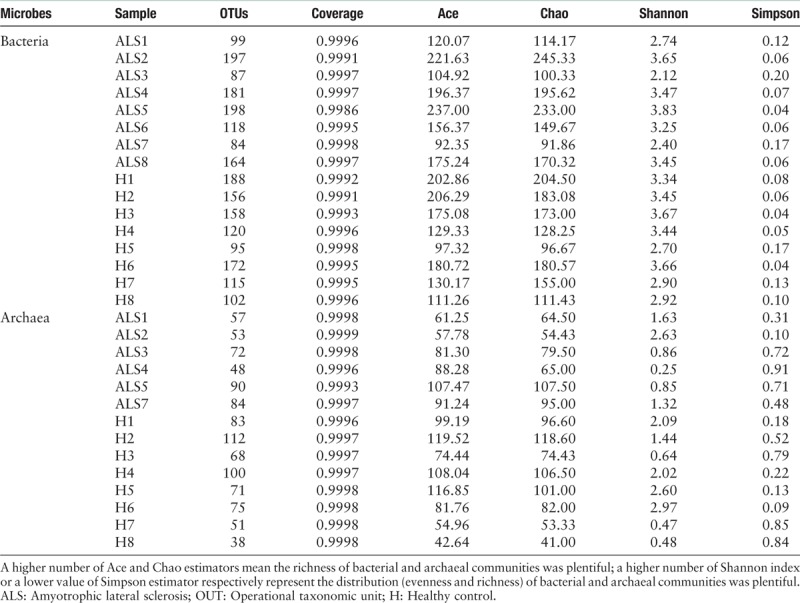
The biodiversity of bacterial and archaeal communities in fecal sample of patients with ALS and healthy individuals were shown in Table 2. The coverage value of each sample maintained approximately 0.9991 to 0.9998. Average OTUs numbers in patients with ALS and healthy individuals were 141 and 138, respectively. Ace and Chao estimators included the richness information of microbial communities. Compared with healthy individuals, the richness of bacterial and archaeal communities in patients with ALS were on the increased and decreased tendencies, respectively. It was generally accepted that the Shannon and Simpson estimators were used to evaluate the richness and evenness of microbial communities. According to these analyses, the richness and evenness of bacterial and archaeal communities of healthy individuals were healthier than those of patients with ALS.
Table 2.
Richness and evenness of micro-organisms in fecal sample of patients with ALS and healthy individuals.
Distribution of micro-organisms at phylum, class, and genus level
As shown in Figure 1A and Supplementary Table S1, bacterial community of fecal sample presented a high predominance of Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia. Notably, the average relative abundance of phylum Firmicutes in patients with ALS was 4.7% higher than that of healthy individuals. On class level, the relative abundance of classes Negativicutes and Bacilli in patients with ALS was on the decline, compared with that of healthy individuals [Figure 1B and Supplementary Table S2]. In Figure 1C and Supplementary Table S3, the top ten genera in patients with ALS and healthy individuals were Bacteroides, Blautia, Faecalibacterium, Escherichia-Shigella, Anaerostipes, Streptococcus, Akkermansia, Fusicatenibacter, Megamonas, and Bifidobacterium (21.1% vs. 25.9%, 15.3% vs. 9.0%, 4.8% vs. 9.9%, 5.9% vs. 3.6%, 4.4% vs. 1.9%, 4.6% vs. 1.0%, 4.5% vs. 0.4%, 1.9% vs. 2.5%, 2.6% vs. 1.4%, and 1.4% vs. 2.5%), which respectively accounted for 65.1% and 55.6% of the total reads in patients with ALS and healthy individuals. In Figure 2 and Supplementary Tables S4 to S6, although the determined archaeal community was under a relatively narrow range, and results indicated that the relative abundance of phylum Euryarchaeota, class Methanobacteria, and genus Methanobrevibacter showed a significant enhancement in patients with ALS, compared with healthy individuals.
Figure 1.
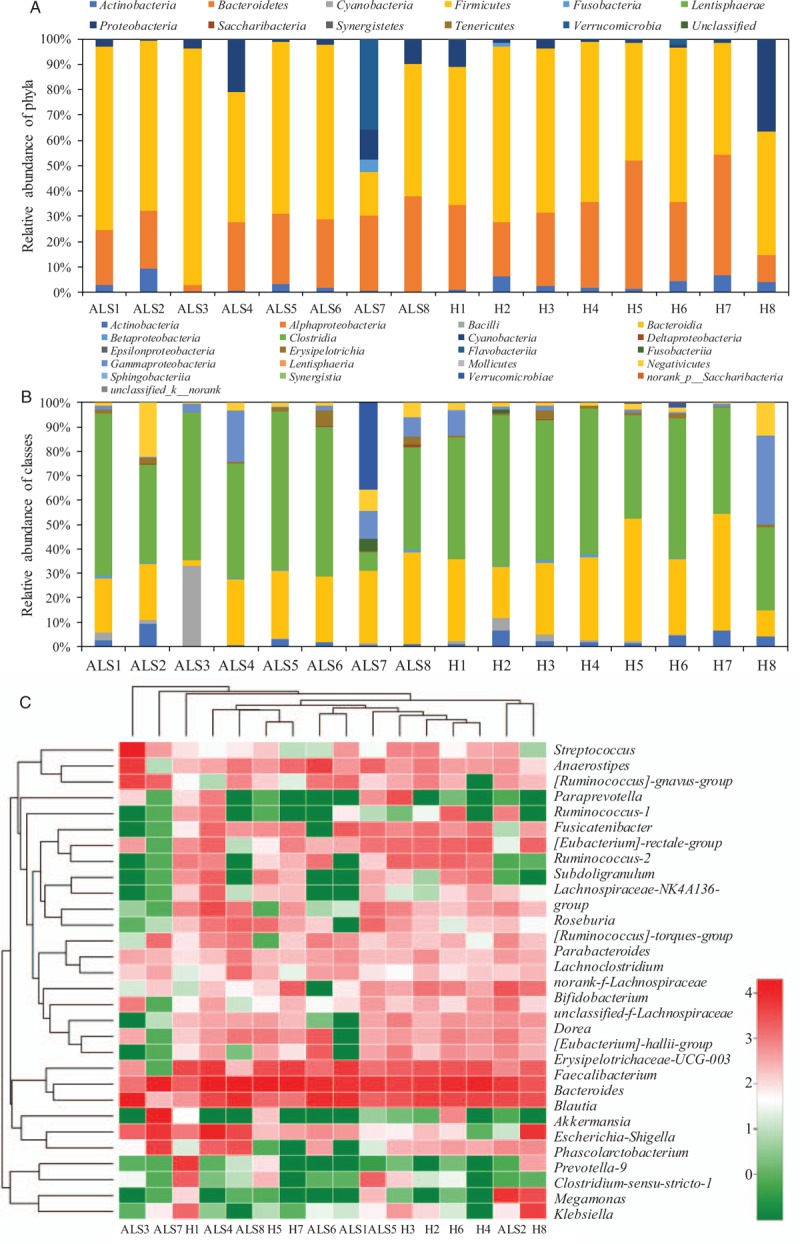
Taxonomic classification of the bacterial communities at phylum (A) and class (B) levels, and hierarchy cluster for the top 30 genera (C) from bacterial throughput sequencing in fecal sample of patients with ALS and healthy individuals. ALS: Amyotrophic lateral sclerosis; H: Healthy control.
Figure 2.
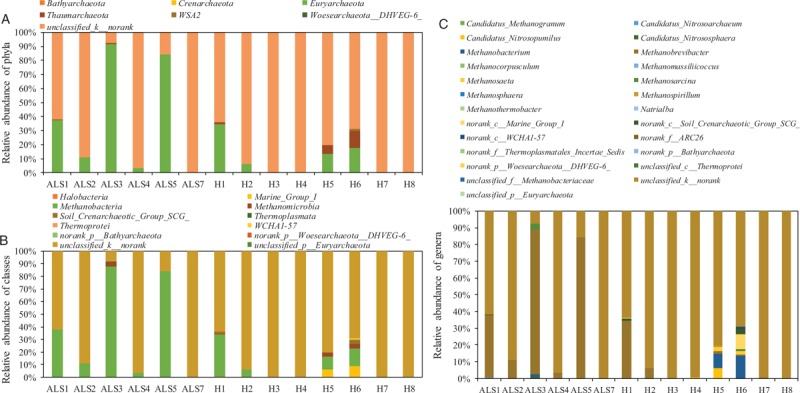
Taxonomic classification of the archaeal communities at phylum (A), class (B), and genus (C) levels from archaeal throughput sequencing in fecal sample of patients with ALS and healthy individuals. ALS: Amyotrophic lateral sclerosis; H: Healthy control.
Metabolite concentration
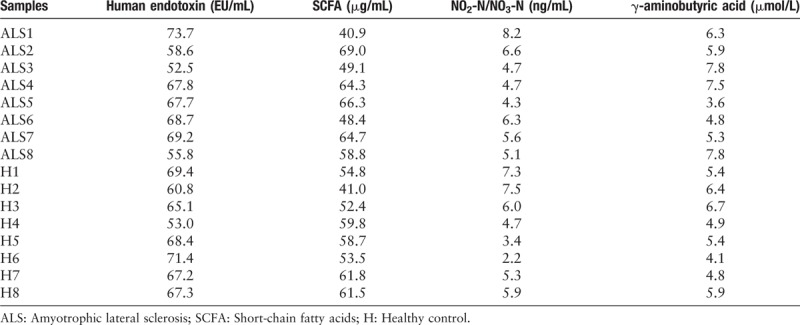
In general, the average concentrations of human endotoxin, SCFA, NO2-N/NO3-N, and γ-aminobutyric acid in patients with ALS and healthy individuals were 64.2 vs. 65.3 EU/mL, 57.5 vs. 55.3 μg/mL, 5.7 vs. 5.3 ng/mL, and 6.1 vs. 5.4 μmol/L, respectively [Table 3]. Although there were no significant differences in these metabolites between patients with ALS and healthy individuals (P = 0.756, 0.614, 0.621, and 0.296, respectively), the results still indicated that the increase trend of SCFA, NO2-N/NO3-N, and γ-aminobutyric acid was observed in patients with ALS, compared with healthy individuals.
Table 3.
Metabolites concentration in fecal sample collected from patients with ALS and healthy individuals.
Discussion
Intestinal microbiome and their interactions in food metabolism play an important role in the nutrient digestion and human health.[24] Most of related studies indicated that the biodiversity, distribution, and metabolism of intestinal microflora showed a significant influence on the healthy and homeostatic balance of human central nervous system.[25–27] Owing to ALS is an age-dependent disease, as the population grows and ages, the incidence population might show an enhancive trend in the world.[4] In addition, ALS is a systemic wasting disease, the patient's weight declined with the digestive disturbances and intestinal barrier dysfunction.[18] Notably, it was observed that the weight of patients with ALS was significantly lower than that of healthy individuals [Table 1]. Therefore, the intestinal flora, which has been considered as the second brain, may be significantly influenced by the ALS disease progressions.[2] Accumulating researches and findings demonstrate that the composition changes of these micro-organisms in the gastrointestinal tract possessed a strongly correlated to the neurological diseases, specifically, neurodegenerative diseases.[2,28]
In the present study, the biodiversity and distribution of intestinal microflora and the metabolite concentrations of these micro-organisms were performed by the high throughput sequencing and the corresponding ELISA kit methods, respectively. Based on the experimental findings, the richness and evenness of bacterial and archaeal communities of healthy individuals were healthier than that of patients with ALS [Table 2]. The increase of bacterial diversity in patients with ALS is caused mainly on the decline of metabolic function, which could have a negative effect on the growth and metabolism of micro-organisms. In literature, it has been indicated that the richness and evenness of bacterial communities showed a decrease trend in patients with ALS or irritable bowel syndrome disease.[2,29,30] In addition, the biodiversity of archaeal communities of patients with ALS was also on the decline compared with that of healthy individuals.
Compared with healthy individuals, the relative abundance of bacterial communities reflected that a higher percent of phylum Firmicutes and a lower percent of phylum Bacteroidetes were found in patients with ALS [Figure 1]. It has been reported that the phylum Firmicutes/Bacteroidetes ratio could be regarded as an important index for the health of human gastrointestinal tract.[2,31] The significantly promoted abundance of phylum Firmicutes and reduced abundance of phylum Bacteroidetes indicated that the gastrointestinal tract health of patients with ALS was significantly influenced by this disease.[2] On genus level, the top ten genera in patients with ALS and healthy individuals were Bacteroides, Blautia, Faecalibacterium, Escherichia-Shigella, Anaerostipes, Streptococcus, Akkermansia, Fusicatenibacter, Megamonas, and Bifidobacterium. The relative abundance of harmful bacteria (genus Dorea) in patients with ALS was observed with an increasing tendency in previous study.[2] However, there were no similar findings obtained in the present study. Notably, compared with the healthy individuals (9.9%), the beneficial micro-organisms (genus Faecalibacterium) was observed with a significant decline in fecal sample of patients with ALS (4.8%).[32,33] In general, this genus maintained an above level of 5% in the gut of healthy people, the decline of genus Faecalibacterium usually represented the development of some diseases like coeliac disease, irritable bowel syndrome, and asthma.[32] In addition, for genus Bacteroides, which could reduce the potential pathogenic factors of gastrointestinal tract in the progression of host intestinal parasitism.[34,35] This genus also presented an apparent decline in fecal sample of patients with ALS.
The mainly determined archaeal communities in fecal sample of patients with ALS and healthy individuals were phylum Euryarchaeota, class Methanobacteria, and genus Methanobrevibacter [Figure 2]. It has been reported that the SCFA could be used as the substrate by genus Methanobrevibacter to produce CH4, meanwhile the weight of host might show a decline trend with the enhancement of this genus.[20] In addition, research on the distribution of archaeal composition indicated that the relative abundance of methanogens in irritable bowel syndrome patients was significantly higher than that of healthy individuals.[16] Similar results were found in fecal sample of patients with ALS compared with healthy individuals. These findings demonstrated that the intestinal metabolic function of patients with ALS might be influenced by this disease.
As shown in Table 3, although there was no significant difference in metabolite concentration between patients with ALS and healthy individuals, the results still indicated an increase trend of SCFA, NO2-N/NO3-N, and γ-aminobutyric acid was observed in patients with ALS compared with healthy individuals. These metabolites have been considered as the important elements for nutrient consumption in daily diet. The concentrations of these metabolites promoted in fecal sample of patients with ALS might demonstrate that the absorptive function of human gastrointestinal tract declined with this disease.
However, there were several limitations in this study. First, no metagenomic sequencing was performed to investigate the genomic information about the microbial function variation in patients with ALS and healthy individuals. Second, to help the patients with ALS establish a healthier gastrointestinal tract, the acclimation of microbial communities might be needed for further studies to have a depth understand about the interactions of intestinal microbiota composition with ALS.
In summary, the biodiversity and composition of intestinal microflora in patients with ALS were generally on the decline compared with healthy individuals. The relative abundance of beneficial micro-organisms (genera Faecalibacterium and Bacteroides) presented with a decrease trend in patients with ALS. In addition, the phylum Firmicutes/Bacteroidetes ratio and genus Methanobrevibacter showed an enhancement trend in patients with ALS indicated that the imbalance of gut flora composition in fecal sample collected from patients with ALS. The concentrations of these metabolites promoted in fecal sample of patients with ALS might demonstrate that the absorptive function of human gastrointestinal tract declined with this disease.
Funding
This study was supported by grants from the Discipline Construction of Important Weak Disciplines in Shanghai Health System (No. 2015ZB0402), Chinese Medicine Research Fund from Shanghai Health and Family Planning Commission (No. 2014LP006A and ZY3-RCPY-4-2005), and Dual Task Training to Prevent Fall of the Elderly Patients with Cognitive (No. SHDCI2014126).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Zhai CD, Zheng JJ, An BC, Huang HF, Tan ZC. Intestinal microbiota composition in patients with amyotrophic lateral sclerosis: establishment of bacterial and archaeal communities analyses. Chin Med J 2019;132:1815–1822. doi: 10.1097/CM9.0000000000000351
References
- 1.Van Es MA, Hardiman O, Chio A, Turner MR, Eisen A, Hardiman O, et al. Amyotrophic lateral sclerosis. Lancet 2017; 390:2084–2098. doi: 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed] [Google Scholar]
- 2.Fang X, Wang X, Yang SG, Meng FJ, Wang XL, Wei H, et al. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol 2016; 7:1479.doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logroscino G, Traynor BJ, Hardiman O, Chiò A, Mitchell D, Swingler RJ, et al. Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry 2010; 81:385–390. doi: 10.1136/jnnp.2009.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YG, Wu SP, Yi JX, Xia YL, Jin DP, Zhou JS, et al. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin Ther 2017; 39:322–336. doi: 10.1016/j.clinthera.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julien JP. Amyotrophic lateral sclerosis: unfolding the toxicity of the misfolded. Cell 2001; 104:581–591. doi: 10.1016/S0092-8674(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 6.Sun QH, Li YR, Lan WJ, Yang F, Cui F, Huang XS. Prognostic value of time to generalization in 71 Chinese patients with sporadic amyotrophic lateral sclerosis. Chin Med J 2019; 132:1023–1027. doi: 101097/CM9.0000000000000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyce PI, Fratta P, Fisher EM, Acevedo-Arozena A. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mamm Genome 2011; 22:420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- 8.Talbot K. Motor neuron disease: the bare essentials. Pract Neurol 2009; 9:303–309. doi: 10.1136/jnnp.2009.188151. [DOI] [PubMed] [Google Scholar]
- 9.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016; 352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015; 350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derrien M, Van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 2015; 23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Erny D, De Angelis ALH, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cani PD, Knauf C. How gut microbes talk to organs: the role of endocrine and nervous routes. Mol Metab 2016; 5:743–752. doi: 10.1016/j.molmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014; 16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol 2011; 11:22.doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attaluri A, Jackson M, Valestin J, Rao SS. Methanogenic flora is associated with altered colonic transit but not stool characteristics in constipation without IBS. Am J Gastroenterol 2010; 105:1407–1411. doi: 10.1038/ajg.2009.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quigley E. The enteric microbiota in the pathogenesis and management of constipation. Best Pract Res Clin Gastroenterol 2011; 25:119–126. doi: 10.1016/j.bpg.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Denise S, José H, Daniel K, Ad AMM, Lennard PLG. Percutaneous endoscopic gastrostomy under conscious sedation in patients with amyotrophic lateral sclerosis is safe. Eur J Gastroenterol Hepatol 2017; 29:1303–1308. doi: 10.1097/MEG.0000000000000959. [DOI] [PubMed] [Google Scholar]
- 19.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004; 4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 20.Dighe AS, Jangid K, González JM, Pidiyar VJ, Patole MS, Ranade DR, et al. Comparison of 16S rRNA gene sequences of genus Methanobrevibacter. BMC Microbiol 2004; 4:20.doi: 10.1186/1471-2180-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000; 1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Xu H, Wang JF, Song XS, Wang YH, Li F, et al. Bacterial and archaeal community distribution and stabilization of anaerobic sludge in a strengthen circulation anaerobic (SCA) reactor for municipal wastewater treatment. Bioresour Technol 2017; 244:750–758. doi: 10.1016/j.biortech.2017.08.040. [DOI] [PubMed] [Google Scholar]
- 23.Davenport M, Poles J, Leung JM, Wolff MJ, Abidi WM, Ullman T, et al. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm Bowel Dis 2014; 20:723–731. doi: 10.1097/MIB.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill JM, Bhattacharjee S, Pogue AI, Lukiw WJ. The gastrointestinal tract microbiome and potential link to Alzheimer's disease. Front Neurol 2014; 5:43.doi: 10.3389/fneur.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grenham S, Clarke G, Cryan JF, Dinan TG. Brain–gut–microbe communication in health and disease. Front Physiol 2011; 2:94.doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins SM, Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology 2009; 136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 27.Sharon G, Sampson TR, Geschwind DH, Sarkis KM. The central nervous system and the gut microbiome. Cell 2016; 167:915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright ML, Fournier C, Houser MC, Malú T, Jonathan G, Vicki SH. Potential role of the gut microbiome in ALS: a systematic review. Biol Res Nurs 2018; 20:513–521. doi: 10.1177/1099800418784202. [DOI] [PubMed] [Google Scholar]
- 29.Ott S, Musfeldt M, Wenderoth D, Hampe J, Brant O, Fölsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009; 9:123.doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 2013; 16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Jiang HY, Ling ZX, Zhang YH, Mao HJ, Ma ZP, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 2015; 48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 2004; 12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Löfmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother 2006; 58:1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


