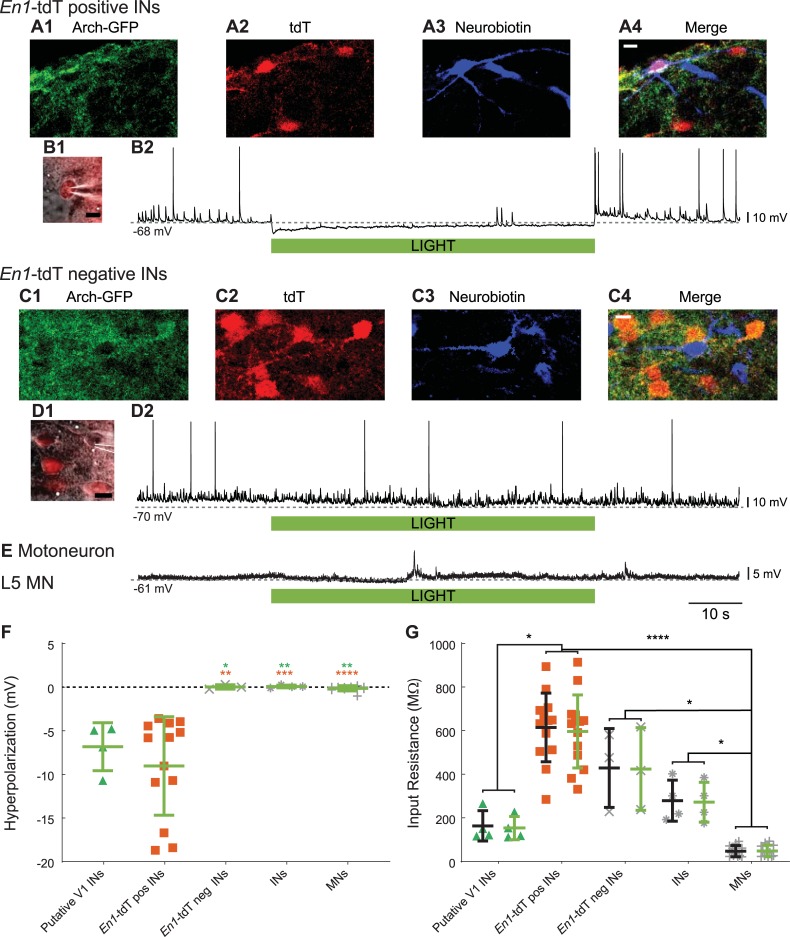
Fig 2. Whole-cell recordings showing the effects of light on V1 INs and MNs in En1-Arch-GFP and En1-Arch-GFP-tdT mouse spinal cords.
(A) Single optical sections (2 μm) of a 100-μm-thick transverse section showing an IN intracellularly filled with Neurobiotin (blue, A3 and A4) expressing both Arch-GFP (green, A1 and A4) and tdT (red, A2 and A4). The white scale bar in A4 is 10 μm. (B1) High-contrast, bright-field image with tdT fluorescence superimposed showing the electrode and the visually targeted tdT-pos IN in a dorsally shaved (between the L2 and L5 ventral roots) En1-Arch-GFP-tdT mouse spinal cord. The black scale bar represents 10 μm. (B2) Intracellular recording of the IN in B1, showing hyperpolarization upon illumination. The duration of the light (60 s) is indicated by the green bar. (C) Single optical sections (2 μm) showing an intracellularly filled tdT-neg IN from a 100-μm-thick transverse section. Panels C1–C4 as in A1–A4. (D1) High-contrast, bright-field image with tdT fluorescence superimposed showing the electrode and a visually targeted tdT-neg IN. The black scale bar represents 10 μm. (D2) Intracellular recording of the IN in D1, showing no hyperpolarization upon illumination. The duration of the light (60 s) is indicated by the green bar. (E) Intracellular recording of a MN recorded in an En1-Arch spinal cord showing no hyperpolarization upon illumination. The duration of the light (60 s) is indicated by the green bar. (F) Plot showing the light-induced hyperpolarization under control conditions for all neurons recorded in this study. All neurons expressing tdT in En1-Arch-GFP-tdT spinal cords were hyperpolarized, whereas neurons neg for tdT were not. MNs did not show any hyperpolarization in En1-Arch-GFP preparations. Unidentified INs in En1-Arch-GFP preparations were classified as putative V1 INs or non-V1 INs according to their response to the light. Both putative V1 INs (green triangles) and En1-tdT-pos INs (red squares) showed a significant hyperpolarization when compared with the other neuronal groups (Kruskall-Wallis test p < 0.0001; putative V1 INs versus En1-tdT-pos INs p = 0.8921, putative V1 INs versus En1-tdT-neg INs p = 0.0224, putative V1 INs versus INs p = 0.0046, putative V1 INs versus MNs p = 0.091; En1-tdT-pos INs versus En1-tdT-neg INs p = 0.0045, En1-tdT-pos INs versus INs p = 0.0003, En1-tdT-pos INs versus MNs p < 0.0001; En1-tdT-neg INs versus INs p = 0.7325, En1-tdT-neg INs versus MNs p = 0.7131; INs versus MNs p = 0.3881; green * represents the p-value comparing putative V1 INs and the other neuronal classes; red * is for the p-value comparing En1-tdT-pos cells and the other neuronal classes). (G) Input resistance of all the INs and MNs recorded in this study under control conditions (black crosses) and during illumination (green crosses), separated according to their putative identities. Putative V1 INs (green triangles; n = 4 in 4 En1-Arch spinal cords, control versus light, Wilcoxon signed-rank test p = 0.375), En1-tdT-pos INs (red rectangles; n = 13 in four En1-Arch-tdT spinal cords, Wilcoxon signed-rank test p = 0.1272), En1-tdT-neg INs (En1-tdT-neg, gray Xs; n = 3 in one En1-Arch-tdT spinal cord, Wilcoxon signed-rank test p > 0.9999), putative non-V1 INs (gray stars; n = 4 in two En1-Arch spinal cords, Wilcoxon signed-rank test p = 0.5), and MNs (gray pluses; n = 12 in 12 En1-Arch spinal cords, Wilcoxon signed-rank test p = 0.0771). Input resistances were also compared among the groups before illumination (Kruskall-Wallis test, p < 0.0001; multiple comparison putative V1 INs versus En1-tdT-pos INs p = 0.0160; putative V1 INs versus En1-tdT-neg INs p = 0.3143; putative V1 INs versus INs p = 0.6182; putative V1 INs versus MNs p = 0.1292; En1-tdT-pos INs versus En1-tdT-neg INs p = 0.3418; En1-tdT-pos INs versus INs p = 0.073; En1-tdT-pos INs versus MNs p < 0.0001; En1-tdT-neg INs versus INs p = 0.5859; En1-tdT-pos INs versus MNs p = 0.0101; INs versus MNs p = 0.0334). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data underlying this figure can be found in S1 Data. Arch, archaerhodopsin-3; En1, engrailed-1; GFP, green fluorescent protein; IN, interneuron; MN, motoneuron; neg, negative; pos, positive; tdT, tdTomato.