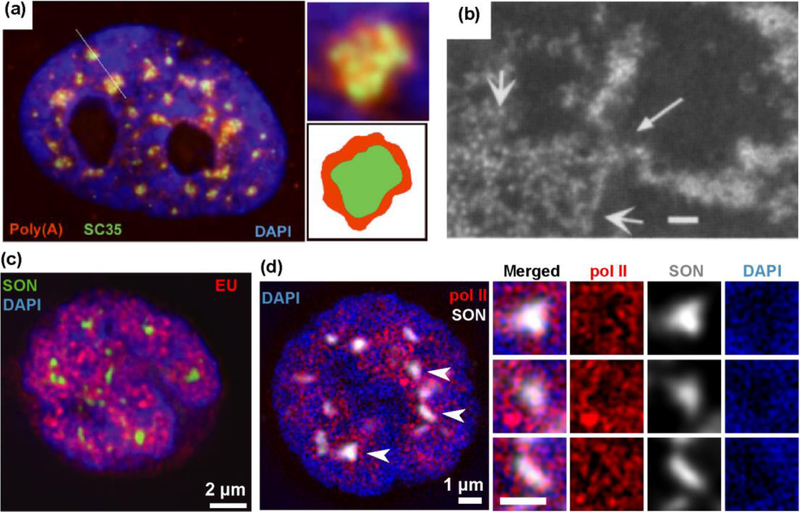
2. Poly(A)+ RNA, Decondensed Chromatin, and Transcription Sites at the Nuclear Speckle Periphery.
(a) Nuclear speckles were identified as “Poly(A)+ RNA islands”(red) and “SC35 domains” (green); the Poly(A)+ RNA signal overlaps but is also peripheral to the SC35 domain. Reprinted from Fig. 2A,E,F, Ref. [2]. (b) Serial-section EM reconstruction of detergent-extracted nucleus stained with heavy metals in CHO cells. Negative image is shown, with heavy metal staining appearing white and low staining black. Locally decondensed large-scale chromatin fibers (small arrowheads) frequently mapped to the Interchromatin Granule Cluster periphery (large arrowheads). Scale bar= 120 nm Reprinted from Fig. 8E, Ref. [8]. (c-e) Active sites of transcription and Ser5p- RNA pol2 staining foci surround nuclear speckle periphery. (d-e) 5 min EU pulse-labeling of transcription (d) or Ser5p-RNA pol 2 staining (e) (red) relative to nuclear speckle marker (SON) (green in (c) and white in (d)). 2x enlarged views corresponding to regions surrounding speckles (arrows). Reprinted and Modified from Fig. 5C&D, Ref. [24, with permission from Rockefeller University Press.