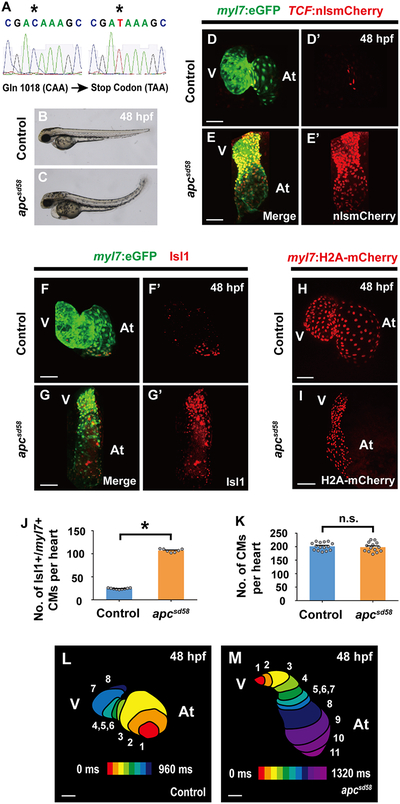
Figure 4. A new apc loss-of-function mutant (apcsd58) displays constitutively activated canonical Wnt signaling and ectopic pacemaker formation.
(A) Sequence analysis of sd58 mutants reveals a C to T nonsense mutation at amino acid 1018 in the apc gene, thus resulting in a putative premature truncation. (B, C) Bright-field microscopy of the lateral side of (B) wild-type (n = 12 embryos) and (C) apcsd58 mutant embryos (n = 10 embryos) shows that apcsd58 mutants display an upwards curved tail and edema at 48 hpf. Confocal images of (D, D’, F, F’, H) wild-type and (E, E’, G, G’, I) apcsd58 mutant embryos at 48 hpf reveal that apcsd58 mutant hearts display (E, E’) increased canonical Wnt-activation as detected by TCF:nlsmCherry (n = 8 embryos) and (G, G’) increased number of Isl1+/myl7+ pacemaker cardiomyocytes (n = 7 embryos) but (I) no appreciable change in overall cardiomyocyte numbers (n = 14 embryos) when compared to (D, D’, F, F’, H) respective wild-type control hearts (n = 7, 9, 15 embryos). Images D’-G’ are single channels from D-G merged images, respectively. Green – (D-G) myl7:eGFP; Red – (D-E’) TCF:nlsmCherry, (F-G’) anti-Isl1 immunostaining, (H, I) myl7:H2A-mCherry. (J, K) Bar graphs show the average number of (J) Isl1+/myl7+ pacemaker cardiomyocytes (CMs) from F-G’ or (K) total cardiomyocytes from H and I. (L, M) Optical mapping of the propagation of calcium activation in (L) control (n = 9 embryos) and (M) apcsd58 mutant (n = 7 embryos) hearts at 48 hpf reveals retrograde cardiac conduction from ventricle to atrium in apcsd58 mutants. Numbers at each isochronal line (60 ms intervals) indicate temporal sequence of calcium activation in the heart. At, atrium; V, ventricle. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. n.s. - not significant. See also Figure S3.