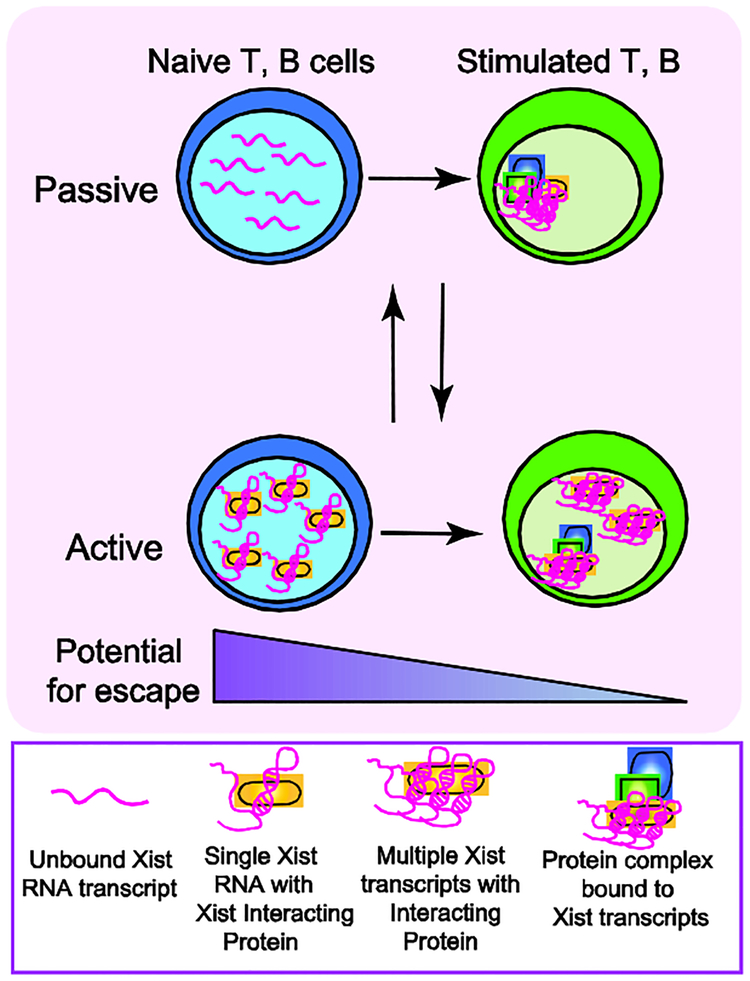
Figure 2: Model for active or passive mechanisms that may regulate dynamic Xist RNA localization during lymphocyte activation.
Mature naïve lymphocytes lack Xist RNA on the Xi, and antigen stimulation triggers Xist RNA return to the Xi. We propose that this re-localization process could occur through either a passive (top) or active (bottom) mechanism. Our passive regulation model posits that unfolded/unstructured Xist RNA transcripts are dispersed across the nucleus, preventing cytological detection. Lymphocyte activation increases levels of Xist RNA interacting proteins, including YY1 and HnRNP-U, which could fold and tether Xist RNA transcripts to the Xi. Alternatively, re-localization could occur via an active process, where in naïve cells we propose that Xist RNA interacting proteins bind and sequester Xist RNA transcripts (perhaps in 1:1 stoichiometry), thereby preventing visualization by RNA FISH. Lymphocyte activation increases the concentrations of Xist RNA interacting proteins, including YY1 and HnRNP-U, which could bind to either one or multiple Xist transcripts. Xist RNA interacting factors then recruit Xist RNA to the Xi and tether Xist RNA transcripts across this chromosome. The active and passive mechanisms are not mutually exclusive. The absence of Xist RNA and heterochromatin marks at the Xi in naive cells may “relax” XCI maintenance and increase the number of genes escaping from XCI, such as Tlr7.