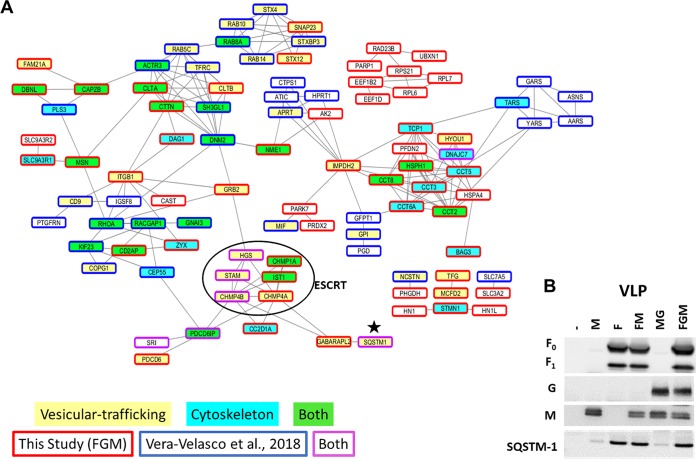
FIG 2.
VLP proteomics highlighted by enrichment of cytoskeletal and endosomal trafficking machinery. (A) STRING protein interaction map comparing proteins identified in the FGM VLPs described by Vera-Velasco et al. (43) (blue box) and this study (red boxes). This map identifies which protein clusters have associated factors included in one or both (purple boxes) studies. Among the most enriched groups based on gene ontological analyses of the combined data set are vesicular trafficking (yellow) and the cytoskeleton (light blue), with proteins that belong to both also designated (green). ESCRTs which somewhat overlap both processes were observed in both studies and are in a black circle. Note that the clusters associated with these processes tend to have included proteins identified in both studies analyzed. Importantly, only proteins with at least one interaction were shown on this map; a full list can be found in Table 1. One of the most clearly enriched proteins in VLP proteomics for this study and that by Vera-Velasco et al. (43) was the autophagy adapter protein, sequestosome-1 (SQSTM1, designated by small black star). (B) Using Western blot analysis of VLPs for each combination, sequestosome-1 was used to validate the accuracy of our VLP proteomics, since this protein exhibited an expression pattern similar to the mass spectrometry results. The Western blot is representative of at least three replicates.