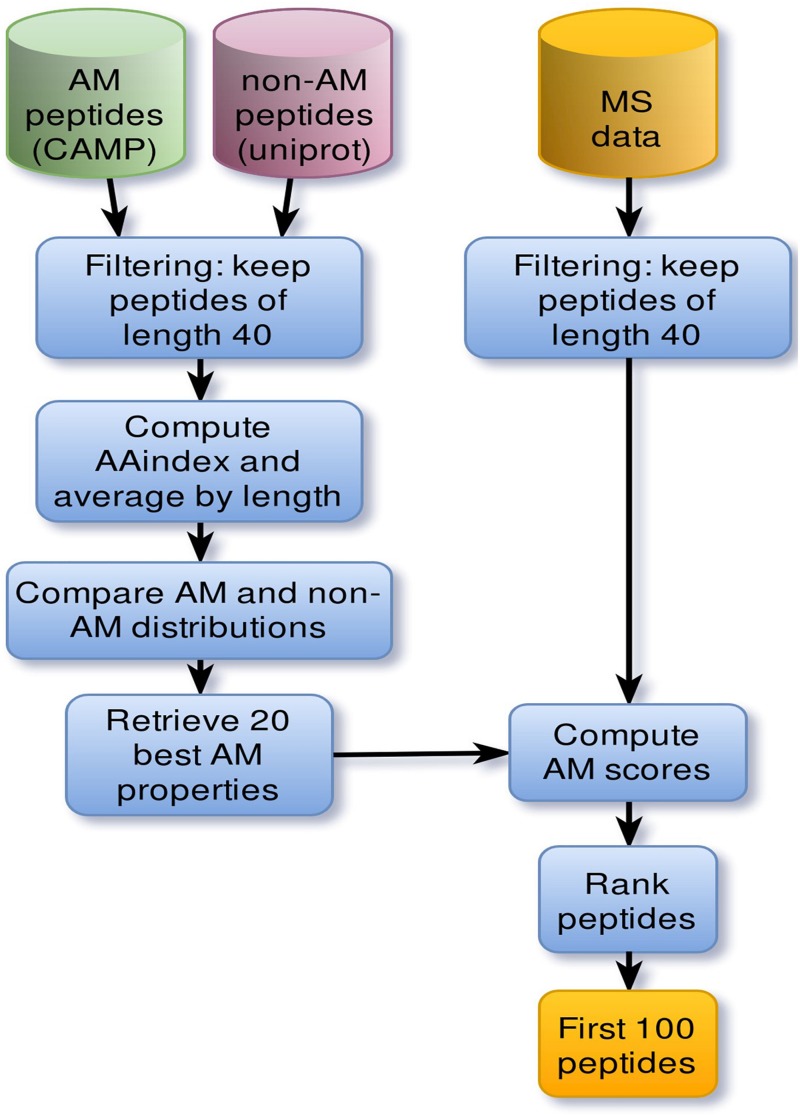
FIGURE 1.
Main steps of the pipeline that led to the discovery of NuriPep 1653. Computational steps involved in the discovery of antimicrobial related properties and AMPs. Cylinders correspond to datasets. First positive (green) and negative (red) datasets were extracted from CAMP and Uniprot databases, respectively. Sequences whose length exceed 40 amino acids were discarded based on the cut-off length for peptides. Sequence physiochemical properties were computed based on individual amino acid scores retrieved from AAindex. Those scores were then normalized by sequence length. Comparison of property distributions yielded 20 main antimicrobial properties which were subsequently used to rank mass spectrometry (MS) peptides. The resulting set of 100 peptides were then manually curated.