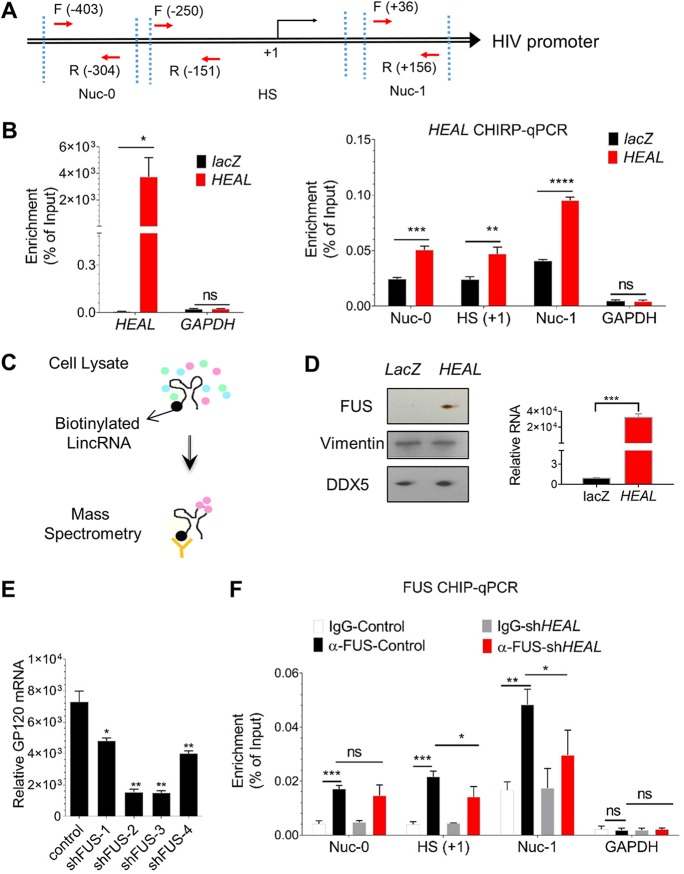
FIG 3.
HEAL forms a complex with FUS protein and binds to HIV promoter. (A) Schematic of HIV promoter regions based on nucleosome architecture. Primers to identify different regions of the promoter are indicated. Nuc-0, nucleosome 0 region. HS, DNase I highly sensitive region. Nuc-1, nucleosome 1 region. (B) HEAL is recruited to the HIV promoter. ChIRP assays were performed in HIV-infected MT4 cells using a nonspecific lacZ probe or HEAL-specific probes. Specificity of HEAL probes (left) and enrichment at the HIV promoter (right) are shown. GAPDH mRNA (left) or genomic GAPDH is shown as negative control. Probes are listed in Table S4. Mean ± SD of n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. (C) Experimental design for purification and identification of HEAL-associated cellular proteins using biotinylated HEAL or lacZ (control) RNA pulldown followed by mass spectrometry. (D) Immunoblotting of FUS, vimentin, and DDX5 proteins identified from biotinylated lacZ and HEAL pulldown assays. Vimentin and DDX5 are shown as negative controls. HEAL mRNA enrichment in pulldown fraction was detected using qPCR. Mean ± SD of n = 3. ***, P < 0.001. (E) FUS knockdown inhibits HIV-1 replication. MT4 cells were infected with control lentivirus (empty vector, pLKO) or lentiviruses carrying FUS-targeting shRNAs. Two days later, they were infected with HIV-1, and GP120 mRNA was quantified by RT-qPCR analysis after 2 days. Signals were normalized to GAPDH mRNA levels. n = 3, mean ± SD; *, P < 0.05; **, P < 0.01. (F) FUS recruitment to the HIV promoter is dependent on HEAL. HIV-infected control or HEAL knockdown MT4 cells were prepared for FUS-CHIP analysis. RT-qPCR of the HIV promoter regions or GAPDH region coimmunoprecipitated with FUS was performed. Mean ± SD of n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001.