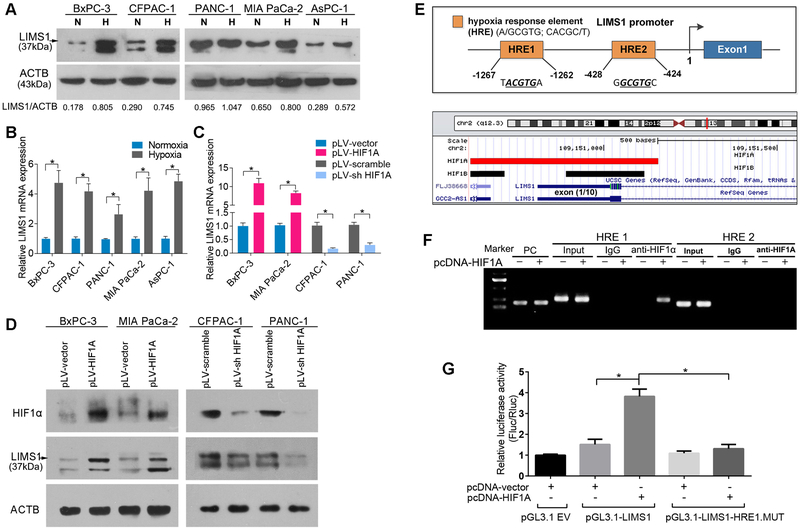
Figure 5. HIF1 transactivates LIMS1 transcription.
A-B, the indicated PDAC tumor cells were incubated in normoxic conditions (N) or hypoxic conditions (H; 1% O2) for 16 hours and subjected to Western blotting (A) and RT-PCR assays (B). C-D, the indicated PDAC tumor cells with HIF1A upregulated or knocked down were subjected to RT-PCR assays (C) and Western blotting (D). E, top: a HIF1 CHIP sequencing data profile (GEO-GSE59937) was downloaded and analysed in UCSC Genome Browser (http://genome.ucsc.edu/). Red rectangle, HIF1A binding site; black rectangle, HIF1β binding site; blue rectangle, the first exon of LIMS1. Bottom: schematic of the human LIMS1 promoter, two hypoxia responsive elements (HREs) were identified (UCSC gene ID: uc002teg.4.). F, CHIP analysis of PANC-1 cells transfected with or without HIF1A-overexpressing plasmids (pcDNA-HIF1A). Chromatin was immunoprecipitated with anti-HIF1A antibody and then subjected to PCR analysis. G, luciferase analysis of Panc-1 cells. Panc-1 cells transfected with pcDNA-HIF1A or control vector (pcDNA-vector) were transfected with pGL3-LIMS1-promoter, pGL3-LIMS1-promoter mutation (MUT), or pGL3-empty vectors (pGL3.1 EV). Forty-eight hours later, cells were subjected to dual luciferase analysis. The results are expressed as a fold induction relative to the cells transfected with the control vector (pcDNA3.1) after normalisation to Renilla activity. Unpaired t tests were used in B, C and G; data are shown as the mean value ± SD; *P < 0.05.