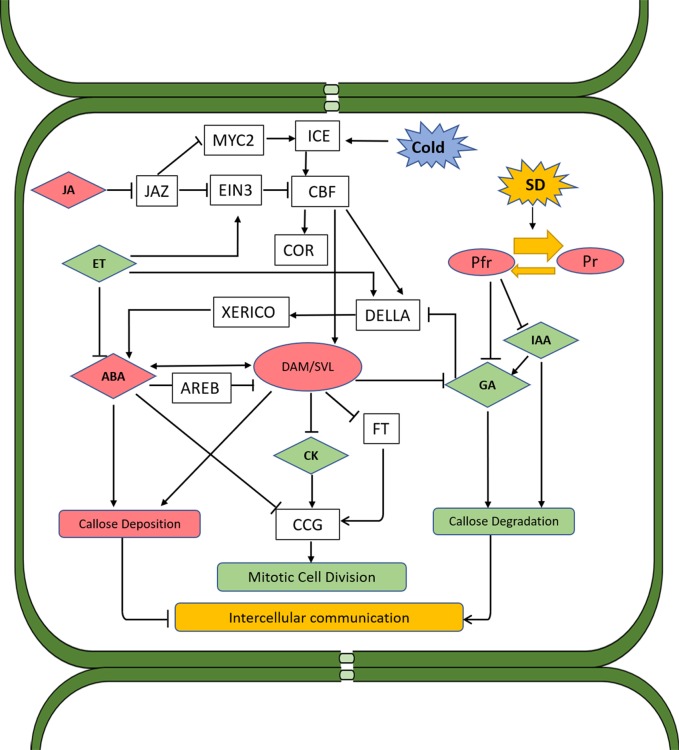
Figure 2.
Proposed schematic model of hormone interactions during bud dormancy induction and release in woody perennials. Solid arrows and lines indicate actions or interactions among hormones, pathways, and environmental cues that have been documented in the literature. Red color indicates the substance or process that induces dormancy, and green color indicates those that promote dormancy release. Both SD photoperiod and low temperature induce CBF, which directly promotes the expression of DAM genes. DAM proteins regulate dormancy through ABA-dependent and ABA-independent pathways. In the former pathway, accumulation of DAMs reduces GA level, thus activating DELLA and subsequently XERICO proteins, which in turn promotes ABA synthesis. DAM genes can also upregulate ABA levels through upregulating the gene encoding NCED, a key enzyme in ABA synthesis. In addition to the control of DAMs over ABA biosynthesis and signaling, ABA can also negatively regulate the expression of DAM genes through SnRK2 and the ABA signaling component AREB. This pathway probably serves as a negative feedback regulation mechanism. In the ABA-independent regulation pathway, DAM proteins induce dormancy through negative regulation of FT, which in turn prevents CCG-mediated dormancy release. ABA can also repress CCG and inhibit the mitotic cell division. ABA was also found to suppress the intercellular communication during dormancy by enhancing the expression of callose synthase, leading to callose deposition and blockage of plasmodesmata. This alteration of plasmodesmata during dormancy is reversed by GA as buds transition to bud break. GAs induce the expression of glucanases which degrade callose, allowing for the passage of sugars and other growth-promoting factors. Under SD conditions, GA biosynthesis is inhibited through phytochrome and phytochrome-interacting factors. ET is induced by SD and has a negative impact on GA biosynthesis and signaling, which nominates it as a dormancy inducer. However, ET also inhibits ABA synthesis and signaling and negatively regulates CBF through activation of EIN3, which is also subject to the regulation of JA through JAZ proteins. JA induces bud dormancy by targeting JAZ proteins for degradation via the ubiquitination/26S proteasome, which in turn releases MYC2 and ICE1 from repression. Both MYC2 and ICE1 activate the expression of CBF. CK can repress ABA and promote dormancy release through inducing the expression of CCGs. IAA facilitates dormancy release through promoting GA biosynthesis and callose degradation. ABA, abscisic acid; CBF, C-repeat binding factor; CCG, cell cycle gene; CK, cytokinin; ET, ethylene; EIN3, ETHYLENE INSENSITIVE 3; FT, flowering locus T; GA, gibberellins; JA, jasmonates; IAA, indole-3-acetic acid; SD, short day; NCED, 9-cis-epoxycarotenoid dioxygenase; DAM, dormancy-associated MADS.