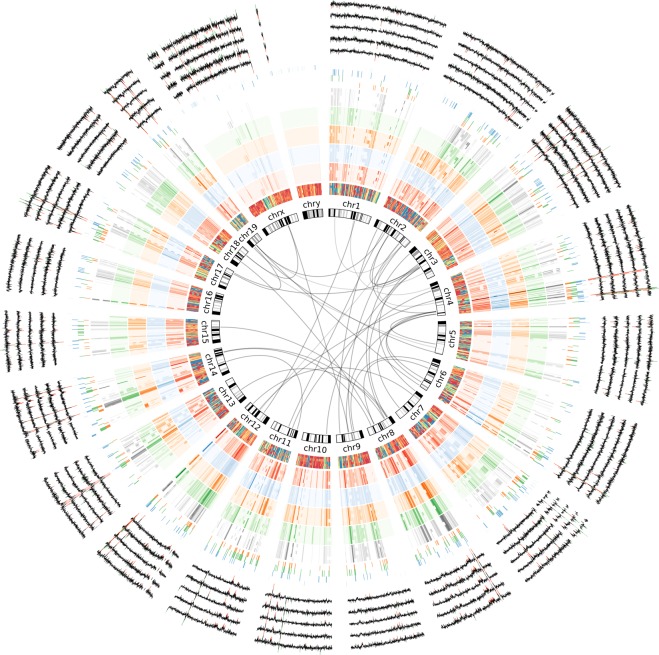
Figure 2.
Circos plot54 showing genetic variation between five individual MutaMouse animals from a colony maintained at Health Canada. Links represent structural translocations identified in all mice sequenced. Each outside track has 5 individual components, one for each mouse sequenced (MutaMouse HC 1, 2, 3, 4, 5, from inside to out). The tracks from inside to outside represent (1) runs of homozygosity (where blue is homozygous and red is heterozygous – immediately adjacent to chromosome numbering), (2) Red track: SNV density per megabase of DNA sequence for all SNVs, (3) Blue track: SNV density for novel SNVs, (4) Orange track: SNV density for variants inherited from DBA/2J exclusively, (5) Green track: SNV density for variants inherited from BALB/cJ exclusively, and (6) Grey track: SNV density for variants inherited from both parents. The next tracks show copy number variants for the parental strains DBA/2J and BALB/cJ (orange and green, respectively), followed by copy number variants in MutaMouse predicted by CNVnator (blue). Finally, the line plots show the log2 values for aCGH probes averaged over 1 megabase intervals, with red highlights showing deleted segments and green highlights showing duplicated segments for each mouse.