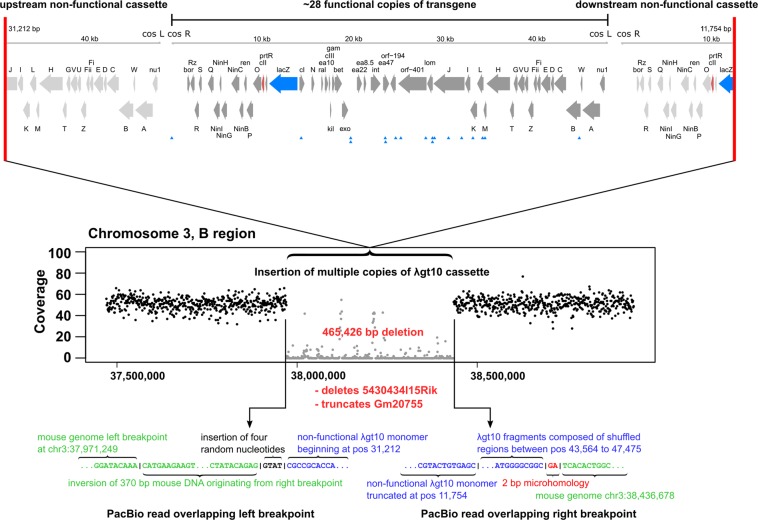
Figure 5.
Assembly of λgt10-lacZ transgene integration site using PacBio read data and NGS coverage data. There are approximately 29 copies of the λgt10 ori replication site35. However, we show here that at least one copy (at the right breakpoint, downstream of the integration site) is non-functional. There are 18 SNVs that are constitutional in every copy of the transgene (shown by blue arrows in the top panel for one transgene monomer, and listed in Table 5). The breakpoints near the integration site were resolved by the use of long PacBio reads that map to both the λgt10 transgene sequence and mouse genomic sequence (bottom panel, colored text). The integration of the transgene comprised an unusual event involving (1) the deletion of ~465 Kbp of mouse genomic sequence, (2) the inversion of 370 bp of mouse genomic sequence from the downstream breakpoint to the upstream breakpoint, (3) the insertion of two partial truncated transgene monomers at either breakpoint, and (4) the insertion of shuffled transgene sequence at the downstream breakpoint as well as the insertion of several random nucleotides at the upstream breakpoint.