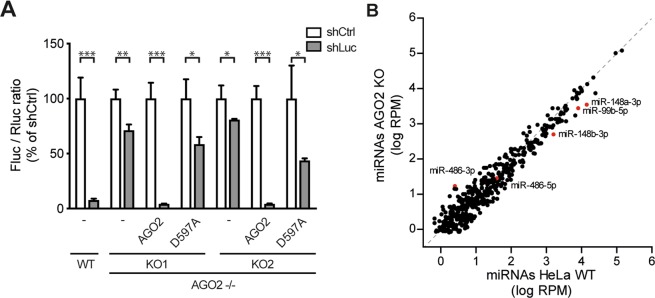
Figure 2.
Functional validation of AGO2 knockout cells. (A) RNAi reporter assay in HeLa WT control and two AGO2 KO cell clones. Cells were transfected with expression plasmids encoding firefly luciferase (Fluc), Renilla luciferase (Rluc), either Fluc specific shRNA (shLuc) or a non-targeting control shRNA (shCtrl), along with expression plasmids encoding wildtype AGO2 or slicer deficient AGO2 D597A where indicated. Fluc counts were normalized to Rluc counts and expressed as a percentage of the non-targeting control shRNA. Data are presented as means and SD of three biological replicates. Data are from one experiment representative of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test. (B) Scatter plot representing miRNA levels in WT and AGO2 KO2 HeLa cells. Reads are normalized to library size. Averaged data from EMCV infected HeLa cells at 8 and 10 hpi are presented (plots for the individual time points in Supplementary Fig. 2). Selected miRNAs are indicated in red.